6TQ6
 
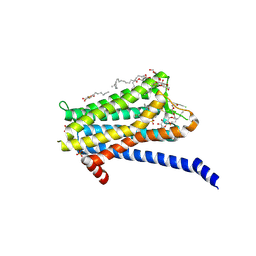 | | Crystal structure of the Orexin-1 receptor in complex with Compound 14 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-(5-methylsulfonylpyridin-3-yl)-1,1-bis(oxidanylidene)-4-[[2,4,6-tris(fluoranyl)phenyl]methyl]pyrido[2,3-e][1,2,4]thiadiazin-3-one, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.546 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6TQI
 
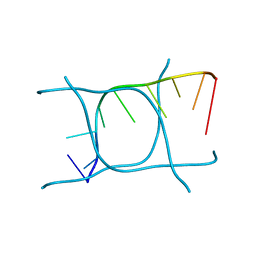 | | I-MOTIF STRUCTURE FORMED FROM THE C STRAND OF A HUMAN TELOMERE FRAGMENT | | Descriptor: | DNA (5'-*TP*AP*AP*CP*CP*CP*TP*AP*A-3') | | Authors: | Parkinson, G.N, Wagner, A, Viladoms-Claverol, J, Duman, R, El-Omari, K. | | Deposit date: | 2019-12-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
6TUI
 
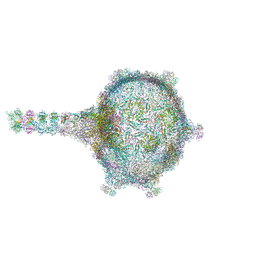 | | Virion of empty GTA particle | | Descriptor: | Adaptor protein Rcc01688, Phage major capsid protein, HK97 family, ... | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2020-01-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (10.47 Å) | | Cite: | Virion of empty GTA particle
Nat Commun, 11, 2020
|
|
5EIT
 
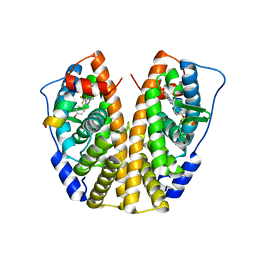 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the imidazopyridine derivative 2-(4-hydroxyphenyl)-3-(trifluoromethyl)imidazo[1,2-a]pyridin-6-ol | | Descriptor: | 2-(4-hydroxyphenyl)-3-(trifluoromethyl)imidazo[1,2-a]pyridin-6-ol, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-10-30 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
6T60
 
 | |
6T6N
 
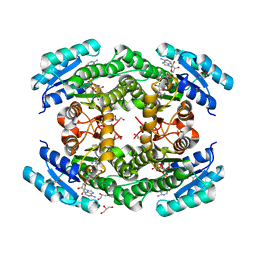 | | Crystal structure of Klebsiella pneumoniae FabG2(NADH-dependent) in complex with NADH at 2.5 A resolution | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-[acyl-carrier protein] reductase, D-MALATE, ... | | Authors: | Vella, P, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens.
Bioorg.Med.Chem., 30, 2021
|
|
5EGH
 
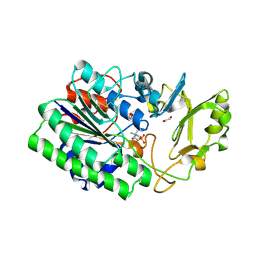 | | Structure of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase in complex with phosphocholine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Morita, J, Kano, K, Kato, K, Takita, H, Ishitani, R, Nishimasu, H, Nureki, O, Aoki, J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structure and biological function of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase
Sci Rep, 6, 2016
|
|
5O64
 
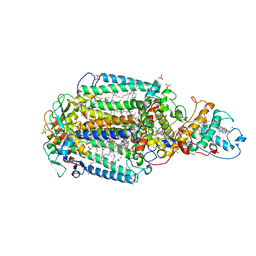 | | From macrocrystals to microcrystals: a strategy for membrane protein serial crystallography | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Dods, R, Baath, P, Branden, G, Neutze, R. | | Deposit date: | 2017-06-05 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | From Macrocrystals to Microcrystals: A Strategy for Membrane Protein Serial Crystallography.
Structure, 25, 2017
|
|
5FQU
 
 | | Orthorhombic crystal structure of of PlpD (selenomethionine derivative) | | Descriptor: | AZIDE ION, PATATIN-LIKE PROTEIN, PLPD, ... | | Authors: | Vinicius da Mata Madeira, P, Zouhir, S, Basso, P, Neves, D, Laubier, A, Salacha, R, Bleves, S, Faudry, E, Contreras-Martel, C, Dessen, A. | | Deposit date: | 2015-12-14 | | Release date: | 2016-04-06 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Basis of Lipid Targeting and Destruction by the Type V Secretion System of Pseudomonas Aeruginosa.
J.Mol.Biol., 428, 2016
|
|
6T86
 
 | | Urocanate reductase in complex with FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Venskutonyte, R, Lindkvist-Petersson, K. | | Deposit date: | 2019-10-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural characterization of the microbial enzyme urocanate reductase mediating imidazole propionate production.
Nat Commun, 12, 2021
|
|
6T87
 
 | | Urocanate reductase in complex with urocanate | | Descriptor: | (2E)-3-(1H-IMIDAZOL-4-YL)ACRYLIC ACID, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Venskutonyte, R, Lindkvist-Petersson, K. | | Deposit date: | 2019-10-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural characterization of the microbial enzyme urocanate reductase mediating imidazole propionate production.
Nat Commun, 12, 2021
|
|
6T88
 
 | |
6T85
 
 | | Urocanate reductase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Venskutonyte, R, Lindkvist-Petersson, K. | | Deposit date: | 2019-10-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural characterization of the microbial enzyme urocanate reductase mediating imidazole propionate production.
Nat Commun, 12, 2021
|
|
5FXG
 
 | | GLUN1B-GLUN2B NMDA RECEPTOR IN ACTIVE CONFORMATION | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
3B6N
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase PV003920 from Plasmodium vivax | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ZINC ION | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase PV003920 from Plasmodium vivax.
To be Published
|
|
5FBQ
 
 | | PI4KB in complex with Rab11 and the MI358 Inhibitor | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta, Ras-related protein Rab-11A, ... | | Authors: | Chalupska, D, Mejdrova, I, Nencka, R, Boura, E. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.789 Å) | | Cite: | PI4KB in complex with Rab11 and the MI358 Inhibitor
To Be Published
|
|
3B6U
 
 | | Crystal structure of the motor domain of human kinesin family member 3B in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF3B, MAGNESIUM ION, ... | | Authors: | Zhu, H, Shen, Y, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Motor domain of human kinesin family member 3B in complex with ADP.
To be Published
|
|
5FES
 
 | | HydE from T. maritima in complex with (2R,4R)-MeSeTDA | | Descriptor: | (2~{R},4~{R})-2-methyl-1,3-selenazolidine-2,4-dicarboxylic acid, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
6TNL
 
 | |
5FF2
 
 | | HydE from T. maritima in complex with (2R,4R)-TDA | | Descriptor: | (2~{R},4~{R})-1,3-thiazolidine-2,4-dicarboxylic acid, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
6TBA
 
 | | Virion of native gene transfer agent (GTA) particle | | Descriptor: | IRON/SULFUR CLUSTER, Phage major capsid protein, HK97 family, ... | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-01 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
5FL5
 
 | | Three dimensional structure of human carbonic anhydrase IX in complex with 5-(1-(4-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)thiophene-2- sulfonamide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-[1-(4-methoxyphenyl)-1,2,3-triazol-4-yl]thiophene-2-sulfonamide, CARBONIC ANHYDRASE IX, ... | | Authors: | Leitans, J, Tars, K, Zalubovskis, R. | | Deposit date: | 2015-10-21 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An Efficient Expression and Crystallization System of the Cancer Asociated Carbonic Anhydrase Isoform Ix.
J.Med.Chem., 58, 2015
|
|
5FGP
 
 | | Crystal structure of D. melanogaster Pur-alpha repeat I-II in complex with DNA. | | Descriptor: | CG1507-PB, isoform B, CHLORIDE ION, ... | | Authors: | Weber, J, Janowski, R, Niessing, D. | | Deposit date: | 2015-12-21 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of nucleic-acid recognition and double-strand unwinding by the essential neuronal protein Pur-alpha.
Elife, 5, 2016
|
|
5FDX
 
 | | Structure of DDR1 receptor tyrosine kinase in complex with D2164 inhibitor at 2.65 Angstroms resolution. | | Descriptor: | 1,2-ETHANEDIOL, 3-[(4-methylpiperazin-1-yl)methyl]-~{N}-[(4~{R})-4-methyl-2-pyrimidin-5-yl-3,4-dihydro-1~{H}-isoquinolin-7-yl]-5-(trifluoromethyl)benzamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bartual, S.G, Pinkas, D.M, Wang, Z, Ding, K, Mahajan, P, Kupinska, K, Mukhopadhyay, S, Strain-Damerell, C, Chalk, R, Borkowska, O, Talon, R, Kopec, J, Williams, E, Tallant, C, Chaikuad, A, Sorell, F, Newman, J, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of DDR1 receptor tyrosine kinase in complex with D2164 inhibitor at 2.65 Angstroms resolution.
To Be Published
|
|
6U0M
 
 | | Structure of the S. cerevisiae replicative helicase CMG in complex with a forked DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA (15-MER), ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Zhang, D, O'Donnell, M, Li, H. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA unwinding mechanism of a eukaryotic replicative CMG helicase.
Nat Commun, 11, 2020
|
|
