5NHY
 
 | | BAY-707 in complex with MTH1 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, GLYCEROL, SULFATE ION, ... | | Authors: | Ellermann, M, Eheim, A, Giese, A, Bunse, S, Nowak-Reppel, K, Neuhaus, R, Weiske, J, Quanz, M, Glasauer, A, Meyer, H, Queisser, N, Irlbacher, H, Bader, B, Rahm, F, Viklund, J, Andersson, M, Ericsson, U, Ginman, T, Forsblom, R, Lindstrom, J, Silvander, C, Tresaugues, L, Gorjanacz, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Novel Class of Potent and Cellularly Active Inhibitors Devalidates MTH1 as Broad-Spectrum Cancer Target.
ACS Chem. Biol., 12, 2017
|
|
5NQ9
 
 | | Crystal structure of laccases from Pycnoporus sanguineus, izoform II, monoclinic | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Orlikowska, M, de J.Rostro-Alanis, M, Bujacz, A, Hernandez-Luna, C, Rubio, R, Parra, R, Bujacz, G. | | Deposit date: | 2017-04-19 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural studies of two thermostable laccases from the white-rot fungus Pycnoporus sanguineus.
Int. J. Biol. Macromol., 107, 2018
|
|
6UG5
 
 | |
6UL4
 
 | |
6OBS
 
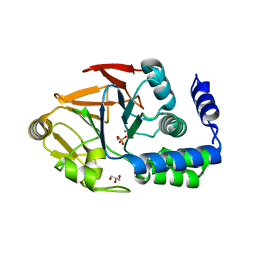 | | PP1 Y134K | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7R7Z
 
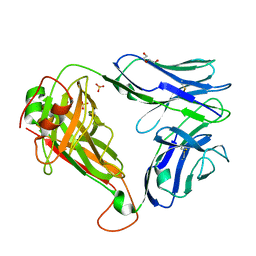 | | Crystal structure of QW9-HLA-B*5301 specific T Cell Receptor, C3 | | Descriptor: | Alpha chain of C3 TCR, Beta chain of C3 TCR, HEXAETHYLENE GLYCOL, ... | | Authors: | Li, X.L, Tan, K.M, Xu, S.T, Ng, R, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.286 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7R7Y
 
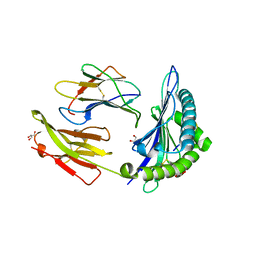 | | Crystal structure of HLA-B*5701 complex with an HIV-1 Gag-derived epitope QW9 S3T variant | | Descriptor: | Beta-2-microglobulin, GLN-ALA-THR-GLN-GLU-VAL-LYS-ASN-TRP, GLYCEROL, ... | | Authors: | Li, X.L, Ng, R, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
5NRA
 
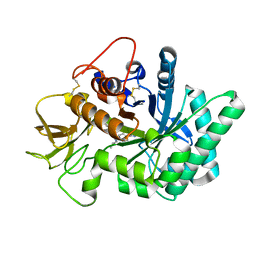 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound 7g | | Descriptor: | 1-(5-azanyl-4~{H}-1,2,4-triazol-3-yl)-~{N}-[2-(4-bromophenyl)ethyl]-~{N}-(2-methylpropyl)piperidin-4-amine, Chitotriosidase-1, GLYCEROL | | Authors: | Mazur, M, Olczak, J, Olejniczak, S, Koralewski, R, Czestkowski, W, Jedrzejczak, A, Golab, J, Dzwonek, K, Dymek, B, Sklepkiewicz, P, Zagozdzon, A, Noonan, T, Mahboubi, K, Conway, B, Sheeler, R, Beckett, P, Hungerford, W.M, Podjarny, A, Mitschler, A, Cousido-Siah, A, Fadel, F, Golebiowski, A. | | Deposit date: | 2017-04-22 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.267 Å) | | Cite: | Targeting Acidic Mammalian chitinase Is Effective in Animal Model of Asthma.
J. Med. Chem., 61, 2018
|
|
6OBU
 
 | | PP1 Y134K in complex with Microcystin LR | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5LW7
 
 | | S. solfataricus ABCE1 post-splitting state | | Descriptor: | ABC transporter ATP-binding protein, IRON/SULFUR CLUSTER | | Authors: | Heuer, A, Gerovac, M, Beckmann, R, Tampe, R. | | Deposit date: | 2016-09-15 | | Release date: | 2016-11-16 | | Last modified: | 2019-02-20 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Structure of the ribosome post-recycling complex probed by chemical cross-linking and mass spectrometry.
Nat Commun, 7, 2016
|
|
5LQW
 
 | | yeast activated spliceosome | | Descriptor: | Pre-mRNA leakage protein 1, Pre-mRNA-processing protein 45, Pre-mRNA-splicing factor 8, ... | | Authors: | Rauhut, R, Luehrmann, R. | | Deposit date: | 2016-08-17 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Molecular architecture of the Saccharomyces cerevisiae activated spliceosome
Science, 6306, 2016
|
|
5VCA
 
 | | VCP like ATPase from T. acidophilum (VAT)-Substrate bound conformation | | Descriptor: | VCP-like ATPase | | Authors: | Ripstein, Z.A, Huang, R, Augustyniak, R, Kay, L.E, Rubinstein, J.L. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-26 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of a AAA+ unfoldase in the process of unfolding substrate.
Elife, 6, 2017
|
|
6QHD
 
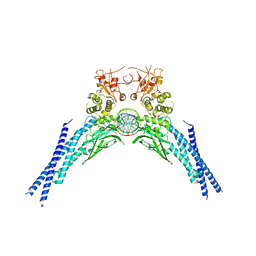 | | Lysine acetylated and tyrosine phosphorylated STAT3 in a complex with DNA | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*TP*TP*TP*AP*CP*GP*GP*GP*AP*AP*AP*TP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*CP*T)-3'), Signal transducer and activator of transcription 3 | | Authors: | Arbely, E, Belo, Y, Shahar, A, Zarivach, R. | | Deposit date: | 2019-01-16 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Unexpected implications of STAT3 acetylation revealed by genetic encoding of acetyl-lysine.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
2BJC
 
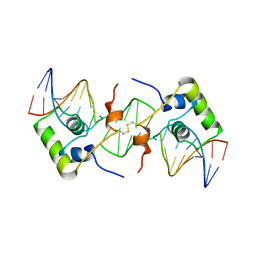 | | NMR structure of a protein-DNA complex of an altered specificity mutant of the lac repressor headpiece that mimics the gal repressor | | Descriptor: | 5'-D(*GP*AP*AP*TP*TP*GP*TP*AP*AP*GP *CP*GP*CP*TP*TP*AP*CP*AP*AP*TP*TP*C)-3', LACTOSE OPERON REPRESSOR | | Authors: | Salinas, R.K, Folkers, G.E, Bonvin, A.M.J.J, Das, D, Boelens, R, Kaptein, R. | | Deposit date: | 2005-02-01 | | Release date: | 2005-10-18 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Altered Specificity in DNA Binding by the Lac Repressor: A Mutant Lac Headpiece that Mimics the Gal Repressor
Chembiochem, 6, 2005
|
|
6VJ3
 
 | | Carbonic Anhydrase II in complex with pyrimidine-based inhibitor | | Descriptor: | 2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-N-(4-sulfamoylphenyl)acetamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Lomelino, C.L, McKenna, R. | | Deposit date: | 2020-01-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Inclusion of a 5-fluorouracil moiety in nitrogenous bases derivatives as human carbonic anhydrase IX and XII inhibitors produced a targeted action against MDA-MB-231 and T47D breast cancer cells.
Eur.J.Med.Chem., 190, 2020
|
|
8AES
 
 | |
5NWY
 
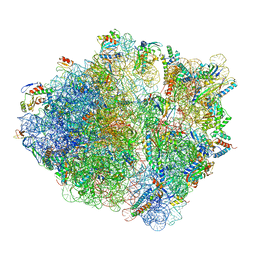 | | 2.9 A cryo-EM structure of VemP-stalled ribosome-nascent chain complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Su, T, Cheng, J, Sohmen, D, Hedman, R, Berninghausen, O, von Heijne, G, Wilson, D.N, Beckmann, R. | | Deposit date: | 2017-05-08 | | Release date: | 2017-07-19 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | The force-sensing peptide VemP employs extreme compaction and secondary structure formation to induce ribosomal stalling.
Elife, 6, 2017
|
|
6QD7
 
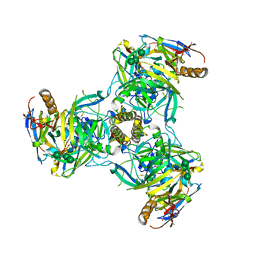 | | EM structure of a EBOV-GP bound to 3T0331 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, Envelope glycoprotein,Virion spike glycoprotein,EBOV-GP1, ... | | Authors: | Diskin, R, Cohen-Dvashi, H. | | Deposit date: | 2019-01-01 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Polyclonal and convergent antibody response to Ebola virus vaccine rVSV-ZEBOV.
Nat. Med., 25, 2019
|
|
7EBT
 
 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in glutathione-bound form | | Descriptor: | CALCIUM ION, GLUTATHIONE, Glutathione transferase | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
7EBW
 
 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in desmethylglycitein and glutathione-bound form | | Descriptor: | 6,7-dihydroxy-3-(4-hydroxyphenyl)-4H-chromen-4-one, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
7EBV
 
 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in luteolin- and glutathione-bound form | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
7EBU
 
 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in Daidzein- and glutathione-bound form | | Descriptor: | 7-hydroxy-3-(4-hydroxyphenyl)-4H-chromen-4-one, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
6WGV
 
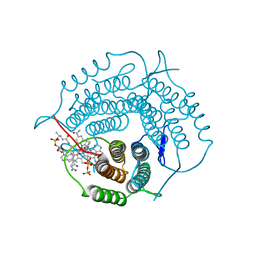 | | Mycobacterium tuberculosis pduO-type ATP:cobalamin adenosyltransferase bound to adenosylcobalamin and PPPi | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Corrinoid adenosyltransferase, ... | | Authors: | Mascarenhas, R.N, Ruetz, M, Koutmos, M, Banerjee, R. | | Deposit date: | 2020-04-06 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Mobile loop dynamics in adenosyltransferase control binding and reactivity of coenzyme B 12 .
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5TDY
 
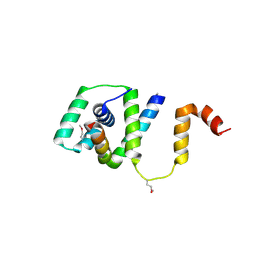 | | Structure of cofolded FliFc:FliGn complex from Thermotoga maritima | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG | | Authors: | Lynch, M.J, Levenson, R, Kim, E.A, Sircar, R, Blair, D.F, Dahlquist, F.W, Crane, B.R. | | Deposit date: | 2016-09-20 | | Release date: | 2017-01-25 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Co-Folding of a FliF-FliG Split Domain Forms the Basis of the MS:C Ring Interface within the Bacterial Flagellar Motor.
Structure, 25, 2017
|
|
6WHC
 
 | | CryoEM Structure of the glucagon receptor with a dual-agonist peptide | | Descriptor: | Dual-agonist peptide, Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Sexton, P, Danev, R. | | Deposit date: | 2020-04-07 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-electron microscopy structure of the glucagon receptor with a dual-agonist peptide.
J.Biol.Chem., 295, 2020
|
|
