8CP4
 
 | | [4Fe-4S] cluster containing LarE in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Zecchin, P, Pecqueur, L, Golinelli-Pimpaneau, B. | | Deposit date: | 2023-03-01 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure-based insights into the mechanism of [4Fe-4S]-dependent sulfur insertase LarE.
Protein Sci., 33, 2024
|
|
4WFS
 
 | | Crystal Structure of tRNA-dihydrouridine(20) synthase catalytic domain | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, SULFATE ION, ... | | Authors: | Bou-Nader, C, Pecqueur, L, Kamah, A, Bregeon, D, Golinelli-Pimpaneau, B, Guimaraes, B.G, Fontecave, M, Hamdane, D. | | Deposit date: | 2014-09-17 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | An extended dsRBD is required for post-transcriptional modification in human tRNAs.
Nucleic Acids Res., 43, 2015
|
|
4WFT
 
 | | Crystal structure of tRNA-dihydrouridine(20) synthase dsRBD domain | | Descriptor: | tRNA-dihydrouridine(20) synthase [NAD(P)+]-like | | Authors: | Bou-Nader, C, Pecqueur, L, Kamah, A, Bregeon, D, Golinelli-Pimpaneau, B, Guimaraes, B.G, Fontecave, M, Hamdane, D. | | Deposit date: | 2014-09-17 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An extended dsRBD is required for post-transcriptional modification in human tRNAs.
Nucleic Acids Res., 43, 2015
|
|
5FRF
 
 | | Solution structure of reduced and zinc-bound RsrA | | Descriptor: | ANTI-SIGMA FACTOR RSRA, ZINC ION | | Authors: | Zdanowski, K, Pecqueur, L, Werner, J, Potts, J.R, Kleanthous, C. | | Deposit date: | 2015-12-17 | | Release date: | 2016-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Anti-Sigma Factor Rsra Responds to Oxidative Stress by Reburying its Hydrophobic Core.
Nat.Commun., 7, 2016
|
|
6EZA
 
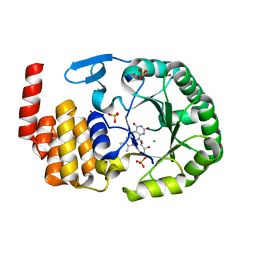 | | Crystal Structure of human tRNA-dihydrouridine(20) synthase catalytic domain E294K mutant | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Bou-Nader, C, Bregeon, D, Pecqueur, L, Vincent, G, Fontecave, M, Hamdane, D. | | Deposit date: | 2017-11-14 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Electrostatic Potential in the tRNA Binding Evolution of Dihydrouridine Synthases.
Biochemistry, 57, 2018
|
|
5FRH
 
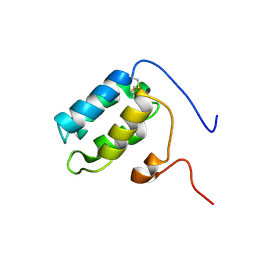 | | Solution structure of oxidised RsrA | | Descriptor: | ANTI-SIGMA FACTOR RSRA | | Authors: | Zdanowski, K, Pecqueur, L, Werner, J, Potts, J.R, Kleanthous, C. | | Deposit date: | 2015-12-17 | | Release date: | 2016-08-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Anti-Sigma Factor Rsra Responds to Oxidative Stress by Reburying its Hydrophobic Core.
Nat.Commun., 7, 2016
|
|
7Z3Z
 
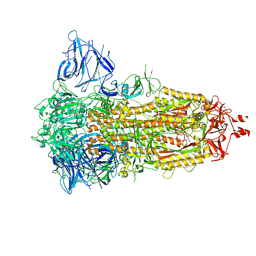 | | Locked Wuhan SARS-CoV2 Prefusion Spike ectodomain with lipid bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, STEARIC ACID, ... | | Authors: | Duyvesteyn, H.M.E, Carrique, L, Ren, J, Stuart, D.I, Fry, E.E. | | Deposit date: | 2022-03-03 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The SARS-CoV-2 Spike harbours a lipid binding pocket which modulates stability of the prefusion trimer
bioRxiv, 2020
|
|
7NDZ
 
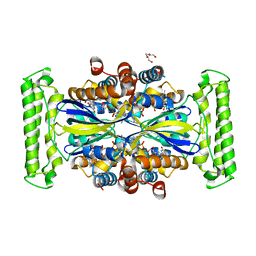 | | ThyX reconstituted with N5-carbinolamine flavin | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent thymidylate synthase, ... | | Authors: | Bou-Nader, C, Pecqueur, L, Hamdane, D. | | Deposit date: | 2021-02-02 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An enzymatic activation of formaldehyde for nucleotide methylation.
Nat Commun, 12, 2021
|
|
7NDW
 
 | | ThyX-FADH2 soaked with 20 mM Formaldehyde | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent thymidylate synthase, ... | | Authors: | Bou-Nader, C, Pecqueur, L, Hamdane, D. | | Deposit date: | 2021-02-02 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An enzymatic activation of formaldehyde for nucleotide methylation.
Nat Commun, 12, 2021
|
|
6Z97
 
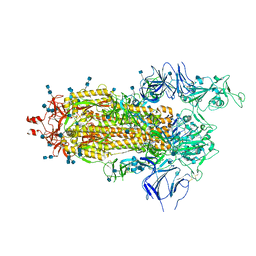 | | Structure of the prefusion SARS-CoV-2 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Duyvesteyn, H.M.E, Ren, J, Zhao, Y, Zhou, D, Huo, J, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-06-03 | | Release date: | 2020-07-01 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6Z43
 
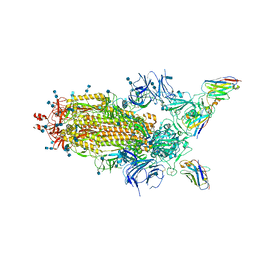 | | Cryo-EM Structure of SARS-CoV-2 Spike : H11-D4 Nanobody Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody, ... | | Authors: | Ruza, R.R, Duyvesteyn, H.M.E, Shah, P, Carrique, L, Ren, J, Malinauskas, T, Zhou, D, Stuart, D.I, Naismith, J.H. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for a potent neutralising single-domain antibody that blocks SARS-CoV-2 binding to its receptor ACE2
To Be Published
|
|
5LAD
 
 | |
6EZB
 
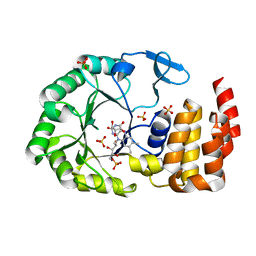 | | Crystal Structure of human tRNA-dihydrouridine(20) synthase catalytic domain Q305K mutant | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Bou-Nader, C, Bregeon, D, Pecqueur, L, Fontecave, M, Hamdane, D. | | Deposit date: | 2017-11-14 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Electrostatic Potential in the tRNA Binding Evolution of Dihydrouridine Synthases.
Biochemistry, 57, 2018
|
|
6EI8
 
 | |
5KH0
 
 | |
6QWL
 
 | | Influenza B virus (B/Panama/45) polymerase Hetermotrimer in complex with 3'5' cRNA promoter | | Descriptor: | 3' cRNA, 5' cRNA, Polymerase acidic protein, ... | | Authors: | Keown, J.R, Carrique, L, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-05 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
8R60
 
 | | 1918 H1N1 Viral polymerase heterotrimer in complex with 4 repeat serine-5 phosphorylated PolII peptide | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2023-11-19 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | 1918 H1N1 Viral polymerase heterotrimer in complex with 4 repeat serine-5 phosphorylated PolII peptide
To Be Published
|
|
8R65
 
 | | 1918 H1N1 Viral polymerase heterotrimer in complex with 4 repeat serine-5 phosphorylated PolII peptide with ordered PB2 C-terminal domains | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2023-11-20 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | 1918 H1N1 Viral polymerase heterotrimer in complex with 4 repeat serine-5 phosphorylated PolII peptide with ordered PB2 C-terminal domains
To Be Published
|
|
7NHX
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer - full transcriptase (Class1) | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NK6
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8204 | | Descriptor: | Nb8204, Polymerase acidic protein, Polymerase basic protein 2,Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-17 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (6.72 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NIS
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8192 core | | Descriptor: | Nanobody8192 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-13 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (5.96 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NK1
 
 | | 1918 Influenza virus polymerase heterotirmer in complex with vRNA promoters and Nb8201 | | Descriptor: | Nanobody8201, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-17 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NJ7
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8200 core | | Descriptor: | Nb8200 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-16 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (4.82 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NKI
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8209 core | | Descriptor: | Nb8209, Polymerase acidic protein, Polymerase basic protein 2,Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-18 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (4.67 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NKA
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8206 | | Descriptor: | NB8206, Polymerase acidic protein, Polymerase basic protein 2,Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-17 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
