4O6W
 
 | | Peptide-Based Inhibitors of Plk1 Polo-box Domain | | Descriptor: | Peptide-Based inhibitor, Serine/threonine-protein kinase PLK1 | | Authors: | Qian, W.-J, Park, J.-E, Lim, D.C, Park, S.-Y, Lee, K.-W, Yaffe, M.B, Lee, K.S, Burke, T.R. | | Deposit date: | 2013-12-23 | | Release date: | 2014-12-03 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Mono-anionic phosphopeptides produced by unexpected histidine alkylation exhibit high plk1 polo-box domain-binding affinities and enhanced antiproliferative effects in hela cells.
Biopolymers, 102, 2014
|
|
6OC9
 
 | | S8 phosphorylated beta amyloid 40 fibrils | | Descriptor: | Amyloid-beta precursor protein, PHOSPHONATE | | Authors: | Qiang, W, Hu, Z.W. | | Deposit date: | 2019-03-22 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Molecular structure of an N-terminal phosphorylated beta-amyloid fibril.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2LNQ
 
 | | 40-residue D23N beta amyloid fibril | | Descriptor: | P3(40) | | Authors: | Qiang, W, Yau, W, Luo, Y, Mattson, M.P, Tycko, R. | | Deposit date: | 2012-01-03 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Antiparallel beta-sheet architecture in Iowa-mutant beta-amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1SK3
 
 | | Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha | | Descriptor: | NICKEL (II) ION, Peptidoglycan recognition protein I-alpha, SULFATE ION | | Authors: | Guan, R, Malchiodi, E.L, Qian, W, Schuck, P, Mariuzza, R.A. | | Deposit date: | 2004-03-04 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha
J.Biol.Chem., 279, 2004
|
|
1SK4
 
 | | crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha | | Descriptor: | Peptidoglycan recognition protein I-alpha, SODIUM ION | | Authors: | Guan, R, Malchiodi, E.L, Qian, W, Schuck, P, Mariuzza, R.A. | | Deposit date: | 2004-03-04 | | Release date: | 2004-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha
J.Biol.Chem., 279, 2004
|
|
4BTL
 
 | | Aromatic interactions in acetylcholinesterase-inhibitor complexes | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Qian, W, Engdahl, C, Berg, L, Ekstrom, F, Linusson, A. | | Deposit date: | 2013-06-18 | | Release date: | 2013-09-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Divergent Structure-Activity Relationships of Structurally Similar Acetylcholinesterase Inhibitors.
J.Med.Chem., 56, 2013
|
|
4DFW
 
 | | Oxime-based Post Solid-phase Peptide Diversification: Identification of High Affinity Polo-like Kinase 1 (Plk1) Polo-box Domain Binding Peptides | | Descriptor: | CHLORIDE ION, Peptide, Serine/threonine-protein kinase PLK1 | | Authors: | Liu, F, Park, J.-E, Qian, W.-J, Lim, D, Gr ber, M, Berg, T, Yaffe, M.B, Lee, K.S, Burke Jr, T.R. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Oxime-based Post Solid-phase Peptide Diversification: Identification of High Affinity Polo-like Kinase 1 (Plk1) Polo-box Domain Binding Peptides
Chem.Biol., 2012
|
|
4X9V
 
 | | PLK-1 polo-box domain in complex with Bioactive Imidazolium-containing phosphopeptide macrocycle 3C | | Descriptor: | Phosphopeptide macrocycle 3C, Serine/threonine-protein kinase PLK1 | | Authors: | Grant, R.A, Qian, W.-J, Yaffe, M.B, Burke, T.R. | | Deposit date: | 2014-12-11 | | Release date: | 2015-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | Neighbor-directed histidine N ( tau )-alkylation: A route to imidazolium-containing phosphopeptide macrocycles.
Biopolymers, 104, 2015
|
|
4X9W
 
 | | PLK-1 polo-box domain in complex with Bioactive Imidazolium-containing phosphopeptide macrocycle 4C | | Descriptor: | Macrocyclic phosphopeptide 4C, Serine/threonine-protein kinase PLK1 | | Authors: | Grant, R.A, Qian, W.-J, Yaffe, M.B, Burke, T.R. | | Deposit date: | 2014-12-11 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Neighbor-directed histidine N ( tau )-alkylation: A route to imidazolium-containing phosphopeptide macrocycles.
Biopolymers, 104, 2015
|
|
4X9R
 
 | | PLK-1 polo-box domain in complex with Bioactive Imidazolium-containing phosphopeptide macrocycle 3B | | Descriptor: | Phosphopeptide macrocycle 3B, Serine/threonine-protein kinase PLK1 | | Authors: | Grant, R.A, Qian, W.-J, Yaffe, M.B, Burke, T.R. | | Deposit date: | 2014-12-11 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Neighbor-directed histidine N ( tau )-alkylation: A route to imidazolium-containing phosphopeptide macrocycles.
Biopolymers, 104, 2015
|
|
3RQ7
 
 | | Polo-like kinase 1 Polo box domain in complex with a C6H5(CH2)8-derivatized peptide inhibitor | | Descriptor: | C6H5(CH2)8-derivatized peptide inhibitor, Serine/threonine-protein kinase PLK1 | | Authors: | Liu, F, Park, J.-E, Qian, W.-J, Lim, D.C, Graber, M, Berg, T, Yaffe, M.B, Lee, K.S, Burke Jr, T.R. | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Serendipitous alkylation of a Plk1 ligand uncovers a new binding channel.
Nat.Chem.Biol., 7, 2011
|
|
2WGU
 
 | | Structure of human adenovirus serotype 37 fibre head in complex with a sialic acid derivative, O-Methyl 5-N- methoxycarbonyl -3,5-dideoxy- D-glycero-a-D-galacto-2-nonulopyranosylonic acid | | Descriptor: | 3,5-dideoxy-5-[(methoxycarbonyl)amino]-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, FIBER PROTEIN, ZINC ION | | Authors: | Johansson, S, Nilsson, E, Qian, W, Guilligay, D, Crepin, T, Cusack, S, Arnberg, N, Elofsson, M. | | Deposit date: | 2009-04-27 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Evaluation of N-Acyl Modified Sialic Acids as Inhibitors of Adenoviruses Causing Epidemic Keratoconjunctivitis.
J.Med.Chem., 52, 2009
|
|
2WGT
 
 | | Structure of human adenovirus serotype 37 fibre head in complex with a sialic acid derivative, O-Methyl 5-N-propaonyl-3,5-dideoxy-D- glycero-a-D-galacto-2-nonulopyranosylonic acid | | Descriptor: | 3,5-dideoxy-5-(propanoylamino)-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, FIBER PROTEIN, ZINC ION | | Authors: | Johansson, S, Nilsson, E, Qian, W, Guilligay, D, Crepin, T, Cusack, S, Arnberg, N, Elofsson, M. | | Deposit date: | 2009-04-27 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Evaluation of N-Acyl Modified Sialic Acids as Inhibitors of Adenoviruses Causing Epidemic Keratoconjunctivitis.
J.Med.Chem., 52, 2009
|
|
3SJO
 
 | | structure of EV71 3C in complex with Rupintrivir (AG7088) | | Descriptor: | 3C protease, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
4B83
 
 | | Mus musculus Acetylcholinesterase in complex with N-(2-Diethylamino- ethyl)-3-methoxy-benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ... | | Authors: | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Berg, L, Qian, W, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-08-24 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Divergent Structure-Activity Relationships of Structurally Similar Acetylcholinesterase Inhibitors.
J.Med.Chem., 56, 2013
|
|
4B80
 
 | | Mus musculus Acetylcholinesterase in complex with N-(2-Diethylamino-ethyl)-1-(4-fluoro-phenyl)-methanesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Berg, L, Qian, W, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-08-24 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Divergent structure-activity relationships of structurally similar acetylcholinesterase inhibitors.
J. Med. Chem., 56, 2013
|
|
3SJ8
 
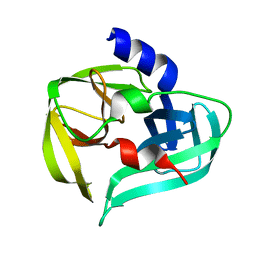 | | Crystal structure of the 3C protease from coxsackievirus A16 | | Descriptor: | 3C protease | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
4ARA
 
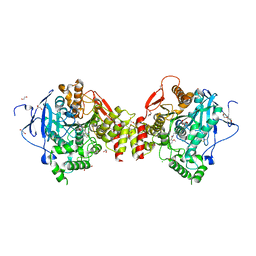 | | Mus musculus Acetylcholinesterase in complex with (R)-C5685 at 2.5 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(DIMETHYLAMINO)-N-{[(2R)-1-ETHYLPYRROLIDIN-2-YL]METHYL}-2-METHOXY-5-NITROBENZAMIDE, ... | | Authors: | Berg, L, Niemiec, M.S, Qian, W, Andersson, C.D, WittungStafshede, P, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-04-23 | | Release date: | 2012-11-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Similar But Different: Thermodynamic and Structural Characterization of a Pair of Enantiomers Binding to Acetylcholinesterase.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3SJI
 
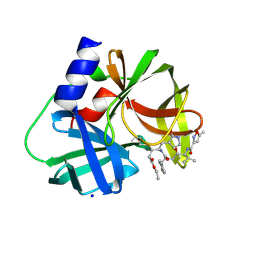 | | crystal structure of CVA16 3C in complex with Rupintrivir (AG7088) | | Descriptor: | 3C protease, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, SODIUM ION | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
4B7Z
 
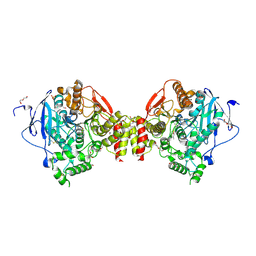 | | Mus musculus Acetylcholinesterase in complex with N-(2-Diethylamino-ethyl)-1-(4-methylphenyl)-methanesulfonamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Berg, L, Qian, W, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-08-24 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Divergent Structure-Activity Relationships of Structurally Similar Acetylcholinesterase Inhibitors.
J.Med.Chem., 56, 2013
|
|
3SJ9
 
 | | crystal structure of the C147A mutant 3C of CVA16 in complex with FAGLRQAVTQ peptide | | Descriptor: | 3C protease, FAGLRQAVTQ peptide | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
3SJK
 
 | | Crystal structure of the C147A mutant 3C from enterovirus 71 | | Descriptor: | 3C protease, KPVLRTATVQGPSLDF peptide | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
4B85
 
 | | Mus musculus Acetylcholinesterase in complex with 4-Chloranyl-N-(2- diethylamino-ethyl)-benzenesulfonamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-chloranyl-N-[2-(diethylamino)ethyl]benzenesulfonamide, ACETYLCHOLINESTERASE, ... | | Authors: | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Berg, L, Qian, W, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-08-24 | | Release date: | 2013-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Divergent Structure-Activity Relationships of Structurally Similar Acetylcholinesterase Inhibitors.
J.Med.Chem., 56, 2013
|
|
4B84
 
 | | Mus musculus Acetylcholinesterase in complex with N-(2-Diethylamino- ethyl)-3-trifluoromethyl-benzenesulfonamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15-PENTAOXAHEPTADECANE, ACETYLCHOLINESTERASE, ... | | Authors: | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Berg, L, Qian, W, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-08-24 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Divergent Structure-Activity Relationships of Structurally Similar Acetylcholinesterase Inhibitors.
J.Med.Chem., 56, 2013
|
|
4B82
 
 | | Mus musculus Acetylcholinesterase in complex with N-(2-Diethylamino- ethyl)-2-fluoranyl-benzenesulfonamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Berg, L, Qian, W, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-08-24 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Divergent Structure-Activity Relationships of Structurally Similar Acetylcholinesterase Inhibitors.
J.Med.Chem., 56, 2013
|
|
