7URN
 
 | |
7URT
 
 | |
8EET
 
 | |
8EEP
 
 | |
8EJL
 
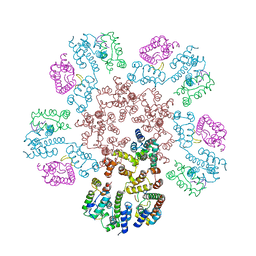 | | Structure of HIV-1 capsid declination in complex with CPSF6-FG peptide | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 6, HIV-1 capsid protein | | Authors: | Pornillos, O, Ganser-Pornillos, B.K, Schirra, R.T, dos Santos, N.F.B. | | Deposit date: | 2022-09-17 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A molecular switch modulates assembly and host factor binding of the HIV-1 capsid.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5I4T
 
 | |
3MGE
 
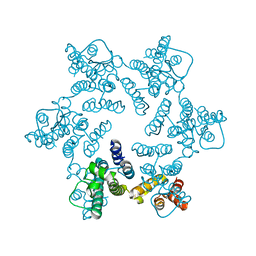 | | X-ray Structure of Hexameric HIV-1 CA | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein p24 | | Authors: | Pornillos, O. | | Deposit date: | 2010-04-05 | | Release date: | 2010-07-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Disulfide Bond Stabilization of the Hexameric Capsomer of Human Immunodeficiency Virus.
J.Mol.Biol., 401, 2010
|
|
4QNB
 
 | |
5TEO
 
 | |
7TGQ
 
 | |
3P0A
 
 | |
3P05
 
 | | X-ray structure of pentameric HIV-1 CA | | Descriptor: | HIV-1 CA, IODIDE ION | | Authors: | Pornillos, O. | | Deposit date: | 2010-09-27 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic-level modelling of the HIV capsid.
Nature, 469, 2011
|
|
7KZH
 
 | |
3H47
 
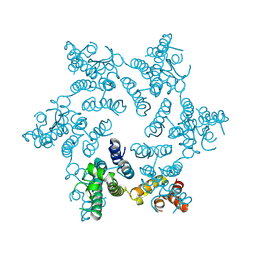 | | X-ray Structure of Hexameric HIV-1 CA | | Descriptor: | Capsid protein p24 | | Authors: | Pornillos, O. | | Deposit date: | 2009-04-18 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of the hexameric building block of the HIV capsid.
Cell(Cambridge,Mass.), 137, 2009
|
|
3H4E
 
 | | X-ray Structure of Hexameric HIV-1 CA | | Descriptor: | Capsid protein p24 | | Authors: | Pornillos, O. | | Deposit date: | 2009-04-19 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | X-ray structures of the hexameric building block of the HIV capsid.
Cell(Cambridge,Mass.), 137, 2009
|
|
4LTB
 
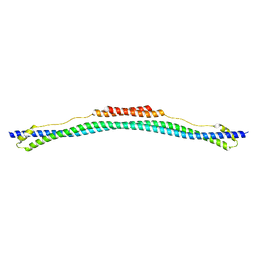 | | Coiled-coil domain of TRIM25 | | Descriptor: | Tripartite motif-containing 25 variant | | Authors: | Pornillos, O, Sanchez, J.G, Okreglicka, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5EYA
 
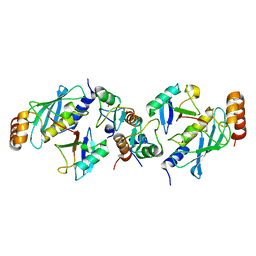 | | TRIM25 RING domain in complex with Ubc13-Ub conjugate | | Descriptor: | Polyubiquitin-B, Tripartite motif-containing 25 variant, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Pornillos, O, Sanchez, J.G. | | Deposit date: | 2015-11-24 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of TRIM25 Catalytic Activation in the Antiviral RIG-I Pathway.
Cell Rep, 16, 2016
|
|
5VZW
 
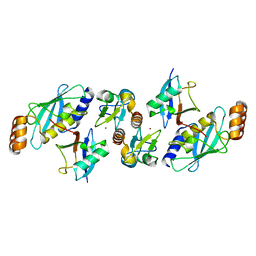 | | TRIM23 RING domain in complex with UbcH5-Ub | | Descriptor: | E3 ubiquitin-protein ligase TRIM23, Ubiquitin, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Pornillos, O, Dawidziak, D. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.278 Å) | | Cite: | Structure and catalytic activation of the TRIM23 RING E3 ubiquitin ligase.
Proteins, 85, 2017
|
|
5VZV
 
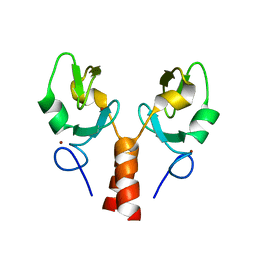 | | TRIM23 RING domain | | Descriptor: | E3 ubiquitin-protein ligase TRIM23, ZINC ION | | Authors: | Pornillos, O, Dawidziak, D. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.812 Å) | | Cite: | Structure and catalytic activation of the TRIM23 RING E3 ubiquitin ligase.
Proteins, 85, 2017
|
|
1KPP
 
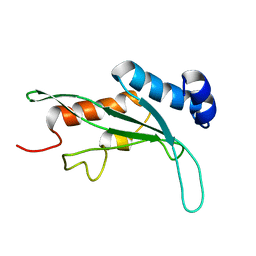 | | Structure of the Tsg101 UEV domain | | Descriptor: | Tumor susceptibility gene 101 protein | | Authors: | Pornillos, O, Alam, S.L, Rich, R.L, Myszka, D.G, Davis, D.R, Sundquist, W.I. | | Deposit date: | 2002-01-02 | | Release date: | 2002-05-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and functional interactions of the Tsg101 UEV domain.
EMBO J., 21, 2002
|
|
1KPQ
 
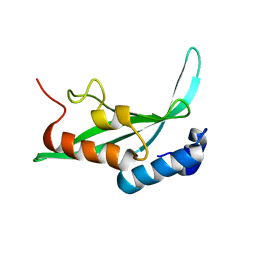 | | Structure of the Tsg101 UEV domain | | Descriptor: | Tumor susceptibility gene 101 protein | | Authors: | Pornillos, O, Alam, S.L, Rich, R.L, Myszka, D.G, Davis, D.R, Sundquist, W.I. | | Deposit date: | 2002-01-02 | | Release date: | 2002-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and functional interactions of the Tsg101 UEV domain.
EMBO J., 21, 2002
|
|
1M4P
 
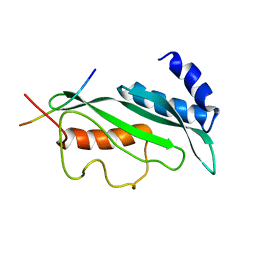 | | Structure of the Tsg101 UEV domain in complex with a HIV-1 PTAP "late domain" peptide, DYANA Ensemble | | Descriptor: | Gag Polyprotein, Tumor Susceptibility gene 101 protein | | Authors: | Pornillos, O, Alam, S.L, Davis, D.R, Sundquist, W.I. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein
Nat.Struct.Biol., 9, 2002
|
|
1M4Q
 
 | | STRUCTURE OF THE TSG101 UEV DOMAIN IN COMPLEX WITH A HIV-1 PTAP "LATE DOMAIN" PEPTIDE, CNS ENSEMBLE | | Descriptor: | Gag Polyprotein, Tumor Susceptibility gene 101 protein | | Authors: | Pornillos, O, Alam, S.L, Davis, D.R, Sundquist, W.I. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein
Nat.Struct.Biol., 9, 2002
|
|
8TY6
 
 | |
7R7P
 
 | | Immature HIV-1 CACTD-SP1 lattice with Bevirimat (BVM) and Inositol hexakisphosphate (IP6) | | Descriptor: | 3alpha-[(3-carboxy-3-methylbutanoyl)oxy]-8alpha,9beta,10alpha,13alpha,17alpha,19beta-lup-20(29)-en-28-oic acid, Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | Sarkar, S, Zadrozny, K.K, Zadorozhnyi, R, Russell, R.W, Quinn, C.M, Kleinpeter, A, Ablan, S, Meshkin, H, Perilla, J.R, Ganser-Pornillos, B.K, Pornillos, O, Freed, E.O, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-06-25 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structural basis of HIV-1 maturation inhibitor binding and activity.
Nat Commun, 14, 2023
|
|
