4PYU
 
 | | The conserved ubiquitin-like protein hub1 plays a critical role in splicing in human cells | | Descriptor: | U4/U6.U5 tri-snRNP-associated protein 1, Ubiquitin-like protein 5 | | Authors: | Ammon, T, Mishra, S.K, Kowalska, K, Popowicz, G.M, Holak, T.A, Jentsch, S. | | Deposit date: | 2014-03-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The conserved ubiquitin-like protein Hub1 plays a critical role in splicing in human cells.
J Mol Cell Biol, 6, 2014
|
|
3JU0
 
 | | Structure of the arm-type binding domain of HAI7 integrase | | Descriptor: | Phage integrase | | Authors: | Szwagierczak, A, Antonenka, U, Popowicz, G.M, Sitar, T, Holak, T.A, Rakin, A. | | Deposit date: | 2009-09-14 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of the arm-type binding domains of HPI and HAI7 integrases
J.Biol.Chem., 284, 2009
|
|
3JTZ
 
 | | Structure of the arm-type binding domain of HPI integrase | | Descriptor: | Integrase, SODIUM ION | | Authors: | Szwagierczak, A, Antonenka, U, Popowicz, G.M, Sitar, T, Holak, T.A, Rakin, A. | | Deposit date: | 2009-09-14 | | Release date: | 2009-10-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the arm-type binding domains of HPI and HAI7 integrases
J.Biol.Chem., 284, 2009
|
|
4QQB
 
 | | Structural basis for the assembly of the SXL-UNR translation regulatory complex | | Descriptor: | Protein sex-lethal, Upstream of N-ras, isoform A, ... | | Authors: | Hennig, J, Popowicz, G.M, Sattler, M. | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the assembly of the Sxl-Unr translation regulatory complex.
Nature, 515, 2014
|
|
3V3B
 
 | | Structure of the Stapled p53 Peptide Bound to Mdm2 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, SAH-p53-8 stapled-peptide | | Authors: | Baek, S, Kutchukian, P.S, Verdine, G.L, Huber, R, Holak, T.A, Ki Won, L, Popowicz, G.M. | | Deposit date: | 2011-12-13 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the stapled p53 peptide bound to Mdm2.
J.Am.Chem.Soc., 134, 2012
|
|
3V3V
 
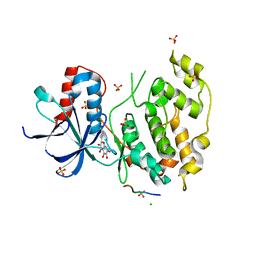 | | Structural and functional analysis of quercetagetin, a natural JNK1 inhibitor | | Descriptor: | 3,5,6,7-TETRAHYDROXY-2-(3,4-DIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, C-Jun-amino-terminal kinase-interacting protein 1, CHLORIDE ION, ... | | Authors: | Baek, S, Kang, N.J, Popowicz, G.M, Arciniega, M, Jung, S.K, Byun, S, Song, N.R, Heo, Y.S, Kim, B.Y, Lee, H.J, Holak, T.A, Augustin, M, Bode, A.M, Huber, R, Dong, Z, Lee, K.W. | | Deposit date: | 2011-12-14 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Analysis of the Natural JNK1 Inhibitor Quercetagetin.
J.Mol.Biol., 425, 2013
|
|
7NDX
 
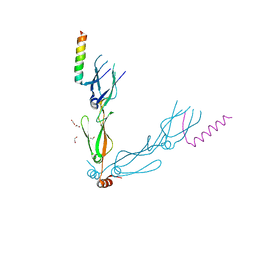 | | Crystal structure of the human HSP40 DNAJB1-CTDs in complex with a peptide of NudC | | Descriptor: | 1,2-ETHANEDIOL, DnaJ homolog subfamily B member 1, Nuclear migration protein nudC | | Authors: | Delhommel, F, Zak, K.M, Popowicz, G.M, Sattler, M. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | NudC guides client transfer between the Hsp40/70 and Hsp90 chaperone systems.
Mol.Cell, 82, 2022
|
|
2GRC
 
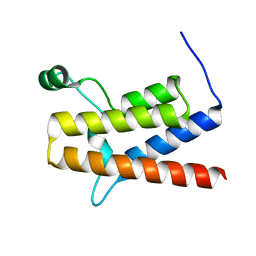 | | 1.5 A structure of bromodomain from human BRG1 protein, a central ATPase of SWI/SNF remodeling complex | | Descriptor: | Probable global transcription activator SNF2L4 | | Authors: | Singh, M, Popowicz, G.M, Krajewski, M, Holak, T.A. | | Deposit date: | 2006-04-24 | | Release date: | 2007-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural ramification for acetyl-lysine recognition by the bromodomain of human BRG1 protein, a central ATPase of the SWI/SNF remodeling complex.
Chembiochem, 8, 2007
|
|
3FDO
 
 | |
3G03
 
 | |
2ABZ
 
 | | Crystal structure of C19A/C43A mutant of leech carboxypeptidase inhibitor in complex with bovine carboxypeptidase A | | Descriptor: | Carboxypeptidase A1, Metallocarboxypeptidase inhibitor, ZINC ION | | Authors: | Arolas, J.L, Popowicz, G.M, Bronsoms, S, Aviles, F.X, Huber, R, Holak, T.A, Ventura, S. | | Deposit date: | 2005-07-18 | | Release date: | 2006-01-31 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Study of a major intermediate in the oxidative folding of leech carboxypeptidase inhibitor: contribution of the fourth disulfide bond
J.Mol.Biol., 352, 2005
|
|
7R2V
 
 | | Structure of nsp14 from SARS-CoV-2 in complex with SAH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Proofreading exoribonuclease nsp14, ... | | Authors: | Czarna, A, Plewka, J, Kresik, L, Matsuda, A, Abdulkarim, K, Robinson, C, OByrne, S, Cunningham, F, Georgiou, I, Pachota, M, Popowicz, G.M, Wyatt, P.G, Dubin, G, Pyrc, K. | | Deposit date: | 2022-02-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Refolding of lid subdomain of SARS-CoV-2 nsp14 upon nsp10 interaction releases exonuclease activity.
Structure, 30, 2022
|
|
3PLU
 
 | | Structure of Hub-1 protein in complex with Snu66 peptide (HINDI) | | Descriptor: | 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, Ubiquitin-like modifier HUB1 | | Authors: | Mishra, S.K, Ammon, T, Popowicz, G.M, Krajewski, M, Nagel, R.J, Ares, M, Holak, T.A, Jentsch, S. | | Deposit date: | 2010-11-15 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing.
Nature, 474, 2011
|
|
6F8G
 
 | |
3PLV
 
 | | Structure of Hub-1 protein in complex with Snu66 peptide (HINDII) | | Descriptor: | 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, Ubiquitin-like modifier HUB1 | | Authors: | Mishra, S.K, Ammon, T, Popowicz, G.M, Krajewski, M, Nagel, R.J, Ares, M, Holak, T.A, Jentsch, S. | | Deposit date: | 2010-11-15 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing.
Nature, 474, 2011
|
|
6F8F
 
 | |
3RMP
 
 | | Structural basis for the recognition of attP substrates by P4-like integrases | | Descriptor: | 5'-D(*TP*AP*AP*TP*GP*AP*CP*CP*AP*CP*CP*AP*AP*TP*A)-3', 5'-D(*TP*AP*TP*TP*GP*GP*TP*GP*GP*TP*CP*AP*TP*TP*A)-3', CP4-like integrase | | Authors: | Szwagierczak, A, Popowicz, G.M, Holak, T.A, Rakin, A, Antonenka, U. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis for the recognition of attP substrates by P4-like integrases
To be Published
|
|
3TJ2
 
 | | Structure of a novel submicromolar MDM2 inhibitor | | Descriptor: | 3-{(1S)-2-(tert-butylamino)-1-[(4-chlorobenzyl)(formyl)amino]-2-oxoethyl}-6-chloro-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, POTASSIUM ION | | Authors: | Wolf, S, Huang, Y, Popowicz, G.M, Goda, S, Holak, T.A, Doemling, A. | | Deposit date: | 2011-08-23 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ugi Multicomponent Reaction Derived p53-Mdm2 Antagonists
To be published
|
|
3UFA
 
 | | Crystal structure of the staphylococcal serine protease SplA in complex with a specific phosphonate inhibitor | | Descriptor: | CHLORIDE ION, N-(3-carboxypropanoyl)-L-valyl-N-[(1S)-2-phenyl-1-phosphonoethyl]-L-prolinamide, Serine protease splA | | Authors: | Zdzalik, M, Pietrusewicz, E, Pustelny, K, Stec-Niemczyk, J, Popowicz, G.M, Potempa, J, Oleksyszyn, J, Dubin, G. | | Deposit date: | 2011-10-31 | | Release date: | 2013-01-23 | | Last modified: | 2014-03-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development and binding characteristics of phosphonate inhibitors of SplA protease from Staphylococcus aureus.
Protein Sci., 23, 2014
|
|
6RZS
 
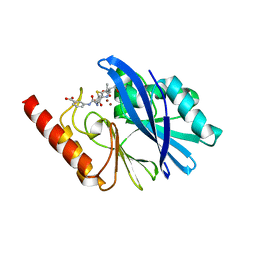 | | Structure of IMP-13 metallo-beta-lactamase complexed with hydrolysed ertapenem | | Descriptor: | Beta-lactamase, ZINC ION, hydrolysed ertapenem | | Authors: | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | Deposit date: | 2019-06-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
6S0H
 
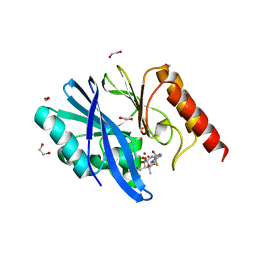 | | Structure of IMP-13 metallo-beta-lactamase complexed with hydrolysed doripenem | | Descriptor: | (2~{R},3~{R})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-3-methyl-4-[(3~{S},5~{S})-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl]sulfanyl-2,3-dihydro-1~{H}-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | Deposit date: | 2019-06-14 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
6RZR
 
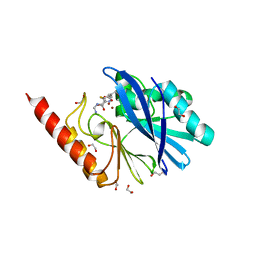 | | Structure of IMP-13 metallo-beta-lactamase complexed with hydrolysed imipenem | | Descriptor: | (2R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-(2-methanimidamidoethylsulfanyl)-2,3-dihydro-1H-pyrrole -5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | Deposit date: | 2019-06-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
6SPT
 
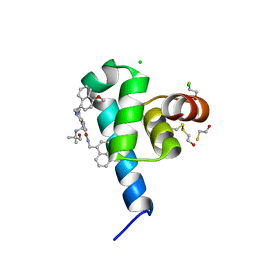 | | High resolution crystal structure of N-terminal domain of PEX14 from Trypanosoma brucei in complex with the fist compound with sub-micromolar trypanocidal activity | | Descriptor: | 5-[(4-methoxynaphthalen-1-yl)methyl]-1-[2-[(2-methyl-1-oxidanyl-propan-2-yl)amino]ethyl]-~{N}-(naphthalen-1-ylmethyl)-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridine-3-carboxamide, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Napolitano, V, Dawidowski, M, Kalel, V.C, Fino, R, Emmanouilidis, L, Lenhart, D, Ostertag, M, Kaiser, M, Kolonko, M, Schilebs, W, Maser, P, Tetko, I, Hadian, K, Plettenburg, O, Erdmann, R, Sattler, M, Popowicz, G.M, Dubin, G. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Activity Relationship in Pyrazolo[4,3-c]pyridines, First Inhibitors of PEX14-PEX5 Protein-Protein Interaction with Trypanocidal Activity.
J.Med.Chem., 63, 2020
|
|
1WQJ
 
 | | Structural Basis for the Regulation of Insulin-Like Growth Factors (IGFs) by IGF Binding Proteins (IGFBPs) | | Descriptor: | Insulin-like growth factor IB, Insulin-like growth factor binding protein 4 | | Authors: | Siwanowicz, I, Popowicz, G.M, Wisniewska, M, Huber, R, Kuenkele, K.P, Lang, K, Engh, R.A, Holak, T.A. | | Deposit date: | 2004-09-29 | | Release date: | 2005-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the regulation of insulin-like growth factors by IGF binding proteins
Structure, 13, 2005
|
|
1ZFI
 
 | | Solution structure of the leech carboxypeptidase inhibitor | | Descriptor: | Metallocarboxypeptidase inhibitor | | Authors: | Arolas, J.L, D'Silva, L, Popowicz, G.M, Aviles, F.X, Holak, T.A, Ventura, S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-09-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structural characterization and computational predictions of the major intermediate in oxidative folding of leech carboxypeptidase inhibitor
STRUCTURE, 13, 2005
|
|
