1M4J
 
 | | CRYSTAL STRUCTURE OF THE N-TERMINAL ADF-H DOMAIN OF MOUSE TWINFILIN ISOFORM-1 | | Descriptor: | A6 gene product | | Authors: | Paavilainen, V.O, Merckel, M.C, Falck, S, Ojala, P.J, Pohl, E, Wilmanns, M, Lappalainen, P. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Conservation Between the Actin Monomer-binding Sites of Twinfilin and Actin-depolymerizing Factor (ADF)/Cofilin
J.Biol.Chem., 277, 2002
|
|
1E3O
 
 | | Crystal structure of Oct-1 POU dimer bound to MORE | | Descriptor: | 5'-D(*AP*TP*GP*CP*AP*TP*GP*AP*GP*GP*A)-3', 5'-D(*TP*CP*CP*TP*CP*AP*TP*GP*CP*AP*T)-3', OCTAMER-BINDING TRANSCRIPTION FACTOR 1 | | Authors: | Remenyi, A, Tomilin, A, Pohl, E, Schoeler, H, Wilmanns, M. | | Deposit date: | 2000-06-20 | | Release date: | 2001-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Dimer Activities of the Transcription Factor Oct-1 by DNA-Induced Interface Swapping
Mol.Cell, 8, 2001
|
|
1FX0
 
 | |
1GU2
 
 | | Crystal structure of oxidized cytochrome c'' from Methylophilus methylotrophus | | Descriptor: | CYTOCHROME C'', HEME C | | Authors: | Enguita, F.J, Pohl, E, Rodrigues, A, Santos, H, Carrondo, M.A. | | Deposit date: | 2002-01-22 | | Release date: | 2003-01-16 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural Evidence for a Proton Transfer Pathway Coupled with Haem Reduction of Cytochrome C" from Methylophilus Methylotrophus.
J.Biol.Inorg.Chem., 11, 2006
|
|
1HF0
 
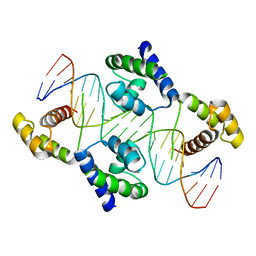 | | Crystal structure of the DNA-binding domain of Oct-1 bound to DNA as a dimer | | Descriptor: | DNA 5'-D(*CP*AP*CP*AP*TP*TP*TP*GP*AP*AP*AP*GP*GP* CP*AP*AP*AP*TP*GP*GP*AP*G)-3', DNA 5'-D(*CP*TP*CP*CP*AP*TP*TP*TP*GP*CP*CP*TP*TP* TP*CP*AP*AP*AP*TP*GP*TP*G)-3', OCTAMER-BINDING TRANSCRIPTION FACTOR 1 | | Authors: | Remenyi, A, Tomilin, A, Pohl, E, Scholer, H.R, Wilmanns, M. | | Deposit date: | 2000-11-27 | | Release date: | 2001-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Differential Dimer Activities of the Transcription Factor Oct-1 by DNA-Induced Interface Swapping
Mol.Cell, 8, 2001
|
|
1BWW
 
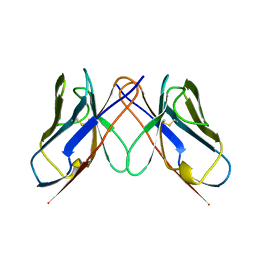 | | BENCE-JONES IMMUNOGLOBULIN REI VARIABLE PORTION, T39K MUTANT | | Descriptor: | PROTEIN (IG KAPPA CHAIN V-I REGION REI) | | Authors: | Uson, I, Pohl, E, Schneider, T.R, Dauter, Z, Schmidt, A, Fritz, H.J, Sheldrick, G.M. | | Deposit date: | 1998-09-29 | | Release date: | 1998-10-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 A structure of the stabilized REIv mutant T39K. Application of local NCS restraints.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
7OB6
 
 | |
7OB7
 
 | |
4CY9
 
 | | DpsA14 from Streptomyces coelicolor | | Descriptor: | DPSA, GLYCEROL | | Authors: | Townsend, P.D, Hitchings, M.D, Del Sol, R, Pohl, E. | | Deposit date: | 2014-04-10 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A Tale of Tails: Deciphering the Contribution of Terminal Tails to the Biochemical Properties of Two Dps Proteins from Streptomyces Coelicolor
Cell.Mol.Life Sci., 71, 2014
|
|
4CYD
 
 | | GlxR bound to cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, GLYCEROL, PROBABLE EXPRESSION TAG, ... | | Authors: | Townsend, P.D, Bott, M, Cann, M.J, Pohl, E. | | Deposit date: | 2014-04-10 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Crystal Structures of Apo and Camp-Bound Glxr from Corynebacterium Glutamicum Reveal Structural and Dynamic Changes Upon Camp Binding in Crp/Fnr Family Transcription Factors.
Plos One, 9, 2014
|
|
4CYB
 
 | | DpsC from Streptomyces coelicolor | | Descriptor: | FE (III) ION, PUTATIVE DNA PROTECTION PROTEIN, SODIUM ION | | Authors: | Townsend, P.D, Hitchings, M.D, Del Sol, R, Pohl, E. | | Deposit date: | 2014-04-10 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A Tale of Tails: Deciphering the Contribution of Terminal Tails to the Biochemical Properties of Two Dps Proteins from Streptomyces Coelicolor
Cell.Mol.Life Sci., 71, 2014
|
|
4CYA
 
 | |
7NGO
 
 | |
7NGG
 
 | |
7NGY
 
 | |
7NGR
 
 | |
7NGI
 
 | |
7NGN
 
 | |
7NGU
 
 | |
7NGD
 
 | |
7NGJ
 
 | |
7NGS
 
 | |
7NGK
 
 | |
7NGT
 
 | |
7NGX
 
 | |
