4ET8
 
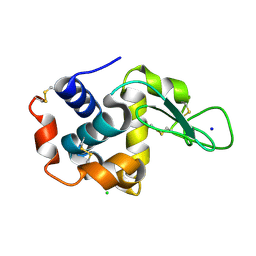 | | Hen egg-white lysozyme solved from 40 fs free-electron laser pulse data | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
8DEZ
 
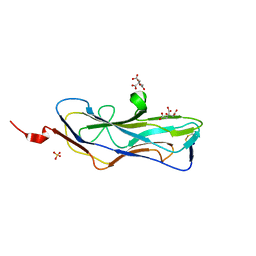 | | Abp2D Receptor Binding Domain ACICU | | Descriptor: | Abp2D Receptor Binding Domain, CITRATE ANION, SULFATE ION | | Authors: | Tamadonfar, K.O, Pinkner, J.P, Dodson, K.W, Kalas, V, Hultgren, S.J. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structure-function correlates of fibrinogen binding by Acinetobacter adhesins critical in catheter-associated urinary tract infections.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DF0
 
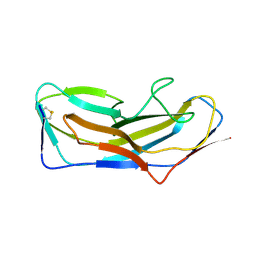 | | Abp1D receptor binding domain | | Descriptor: | Abp1D Receptor Binding Domain | | Authors: | Tamadonfar, K.O, Pinkner, J.S, Dodson, K.W, Hultgren, S.J. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-function correlates of fibrinogen binding by Acinetobacter adhesins critical in catheter-associated urinary tract infections.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DKA
 
 | |
5U91
 
 | | Crystal structure of Tre/loxLTR complex | | Descriptor: | DNA (37-MER), Tre recombinase protein | | Authors: | Meinke, G, Karpinski, J, Buchholz, F, Bohm, A. | | Deposit date: | 2016-12-15 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Crystal structure of an engineered, HIV-specific recombinase for removal of integrated proviral DNA.
Nucleic Acids Res., 45, 2017
|
|
6ALT
 
 | | Solution structure of a DNA dodecamer with 5-methylcytosine at the 3rd and 9th position | | Descriptor: | DNA (5'-D(*(DC5)P*GP*(DMC)P*GP*AP*AP*TP*TP*(DMC)P*GP*CP*(DG3))-3') | | Authors: | Gruber, D.R, Hoppins, J.J, Miears, H.L, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
1TR7
 
 | | FimH adhesin receptor binding domain from uropathogenic E. coli | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, FimH protein, ... | | Authors: | Bouckaert, J, Berglund, J, Schembri, M, De Genst, E, Cools, L, Wuhrer, M, Hung, C.S, Pinkner, J, Slattegard, R, Zavialov, A, Choudhury, D, Langermann, S, Hultgren, S.J, Wyns, L, Klemm, P, Oscarson, S, Knight, S.D, De Greve, H. | | Deposit date: | 2004-06-21 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Receptor binding studies disclose a novel class of high-affinity inhibitors of the Escherichia coli FimH adhesin
Mol.Microbiol., 55, 2005
|
|
8DI2
 
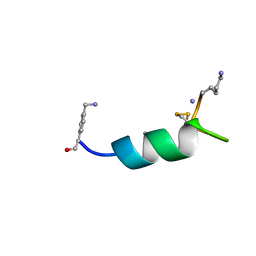 | |
5YVS
 
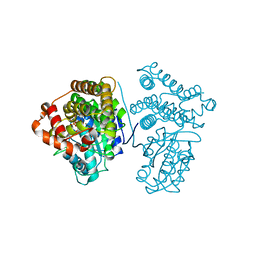 | | Crystal Structure of the archaeal halo-thermophilic Red Sea brine pool alcohol dehydrogenase ADH/D1 bound to NADP | | Descriptor: | MANGANESE (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, alcohol dehydrogenase | | Authors: | Groetzinger, S.W, Strillinger, E, Frank, A, Eppinger, J, Groll, M, Arold, S.T. | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.345 Å) | | Cite: | Identification and Experimental Characterization of an Extremophilic Brine Pool Alcohol Dehydrogenase from Single Amplified Genomes
ACS Chem. Biol., 13, 2018
|
|
5YVR
 
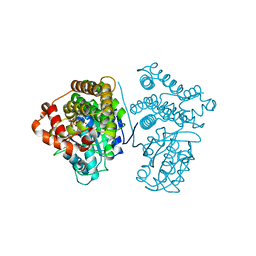 | | Crystal Structure of the H277A mutant of ADH/D1, an archaeal halo-thermophilic Red Sea brine pool alcohol dehydrogenase | | Descriptor: | MANGANESE (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, alcohol dehydrogenase | | Authors: | Groetzinger, S.W, Strillinger, E, Frank, A, Eppinger, J, Groll, M, Arold, S.T. | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Identification and Experimental Characterization of an Extremophilic Brine Pool Alcohol Dehydrogenase from Single Amplified Genomes
ACS Chem. Biol., 13, 2018
|
|
5YVM
 
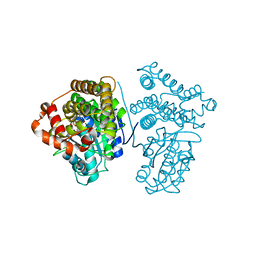 | | Crystal Structure of the archaeal halo-thermophilic Red Sea brine pool alcohol dehydrogenase ADH/D1 bound to NZQ | | Descriptor: | 5,6-DIHYDROXY-NADP, MANGANESE (II) ION, alcohol dehydrogenase | | Authors: | Groetzinger, S.W, Strillinger, E, Frank, A, Eppinger, J, Groll, M, Arold, S.T. | | Deposit date: | 2017-11-26 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Identification and Experimental Characterization of an Extremophilic Brine Pool Alcohol Dehydrogenase from Single Amplified Genomes
ACS Chem. Biol., 13, 2018
|
|
1ZEO
 
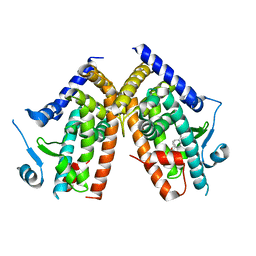 | | Crystal Structure of Human PPAR-gamma Ligand Binding Domain Complexed with an Alpha-Aryloxyphenylacetic Acid Agonist | | Descriptor: | (2S)-(4-ISOPROPYLPHENYL)[(2-METHYL-3-OXO-5,7-DIPROPYL-2,3-DIHYDRO-1,2-BENZISOXAZOL-6-YL)OXY]ACETATE, Peroxisome proliferator activated receptor gamma | | Authors: | Shi, G.Q, Dropinski, J.F, McKeever, B.M, Adams, A.D, MacNaul, K.L, Elbrecht, A, Berger, J.P, Zhou, G, Doebber, T.W. | | Deposit date: | 2005-04-19 | | Release date: | 2006-04-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and Synthesis of alpha-Aryloxyphenylacetic Acid Derivatives: A Novel Class of PPAR alpha/gamma Dual Agonists with Potent Antihyperglycemic and Lipid Modulating Activity
J.Med.Chem., 48, 2005
|
|
4AAB
 
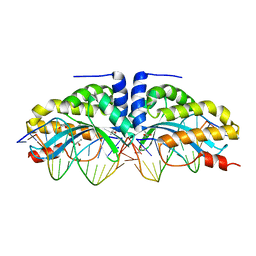 | | Crystal structure of the mutant D75N I-CreI in complex with its wild- type target (The four central bases, 2NN region, are composed by GTAC from 5' to 3') | | Descriptor: | 10MER DNA 5'-D(*GP*AP*CP*GP*TP*TP*TP*TP*GP*AP)-3', 14MER DNA 5'-D(*TP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*AP*CP)-3', DNA ENDONUCLEASE I-CREI, ... | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F.L, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
4AAD
 
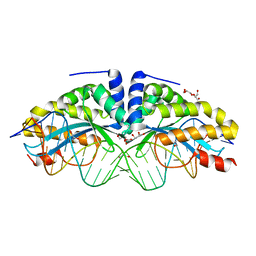 | | Crystal structure of the mutant D75N I-CreI in complex with its wild- type target in absence of metal ions at the active site (The four central bases, 2NN region, are composed by GTAC from 5' to 3') | | Descriptor: | 24MER DNA, DNA ENDONUCLEASE I-CREI, GLYCEROL | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F.L, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
4AAG
 
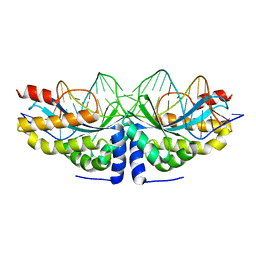 | | Crystal structure of the mutant D75N I-CreI in complex with its wild- type target in presence of Ca at the active site (The four central bases, 2NN region, are composed by GTAC from 5' to 3') | | Descriptor: | 5'-D(*TP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*AP*CP *GP*AP*CP*GP*TP*TP*TP*TP*GP*A)-3', CALCIUM ION, DNA ENDONUCLEASE I-CREI | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
3DCV
 
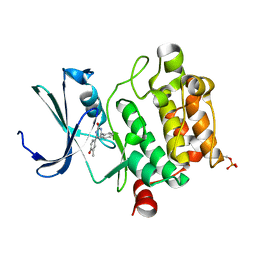 | | Crystal structure of human Pim1 kinase complexed with 4-(4-hydroxy-3-methyl-phenyl)-6-phenylpyrimidin-2(1H)-one | | Descriptor: | 4-(4-hydroxy-3-methylphenyl)-6-phenylpyrimidin-2(5H)-one, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Bellamacina, C.R, Shafer, C.M, Lindvall, M, Gesner, T.G, Yabannavar, A, Weiping, J, Song, L, Walter, A. | | Deposit date: | 2008-06-04 | | Release date: | 2008-08-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 4-(1H-indazol-5-yl)-6-phenylpyrimidin-2(1H)-one analogs as potent CDC7 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4EUL
 
 | | Crystal structure of enhanced Green Fluorescent Protein to 1.35A resolution reveals alternative conformations for Glu222 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Green fluorescent protein, ... | | Authors: | Jones, D.D, Arpino, J.A.J, Rizkallah, P.J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of enhanced green fluorescent protein to 1.35 a resolution reveals alternative conformations for glu222.
Plos One, 7, 2012
|
|
4AAE
 
 | | Crystal structure of the mutant D75N I-CreI in complex with an altered target (The four central bases, 2NN region, are composed by AGCG from 5' to 3') | | Descriptor: | 24MER DNA, DNA ENDONUCLEASE I-CREI | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F.L, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
4AAF
 
 | | Crystal structure of the mutant D75N I-CreI in complex with an altered target (The four central bases, 2NN region, are composed by TGCA from 5' to 3') | | Descriptor: | 1,2-ETHANEDIOL, 24MER DNA, DNA ENDONUCLEASE I-CREI | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F.L, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
4BWT
 
 | | Three-dimensional structure of Paracoccus pantotrophus pseudoazurin at pH 6.5 | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN, SULFATE ION | | Authors: | Freire, F, Mestre, A, Pinho, J, Najmudin, S, Bonifacio, C, Pauleta, S.R, Romao, M.J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Exploring the Surface Determinants of Paracoccus Pantotrophus Pseudoazurin
To be Published
|
|
4BXV
 
 | | Three-dimensional structure of the mutant K109A of Paracoccus pantotrophus pseudoazurin at pH 7.0 | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN, SULFATE ION | | Authors: | Freire, F, Mestre, A, Pinho, J, Najmudin, S, Bonifacio, C, Pauleta, S.R, Romao, M.J. | | Deposit date: | 2013-07-15 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Exploring the Surface Determinants of Paracoccus Pantotrophus Pseudoazurin
To be Published
|
|
4BWU
 
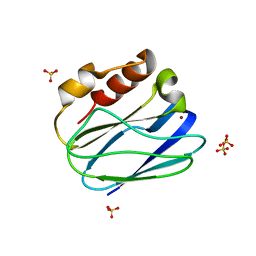 | | Three-dimensional structure of the K109A mutant of Paracoccus pantotrophus pseudoazurin at pH 5.5 | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN, SULFATE ION | | Authors: | Freire, F, Mestre, A, Pinho, J, Najmudin, S, Bonifacio, C, Pauleta, S.R, Romao, M.J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Exploring the Surface Determinants of Paracoccus Pantotrophus Pseudoazurin
To be Published
|
|
1QUN
 
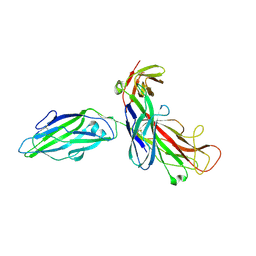 | | X-RAY STRUCTURE OF THE FIMC-FIMH CHAPERONE ADHESIN COMPLEX FROM UROPATHOGENIC E.COLI | | Descriptor: | MANNOSE-SPECIFIC ADHESIN FIMH, PAPD-LIKE CHAPERONE FIMC | | Authors: | Choudhury, D, Thompson, A, Stojanoff, V, Langerman, S, Pinkner, J, Hultgren, S.J, Knight, S. | | Deposit date: | 1999-07-01 | | Release date: | 1999-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli.
Science, 285, 1999
|
|
1YDX
 
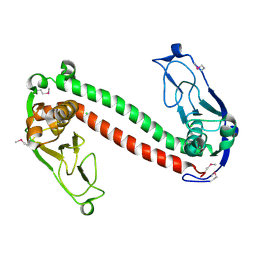 | | Crystal structure of Type-I restriction-modification system S subunit from M. genitalium | | Descriptor: | CHLORIDE ION, type I restriction enzyme specificity protein MG438 | | Authors: | Machado, B, Quijada, O, Pinol, J, Fita, I, Querol, E, Carpena, X. | | Deposit date: | 2004-12-27 | | Release date: | 2005-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Putative Type I Restriction-Modification S Subunit from Mycoplasma genitalium
J.Mol.Biol., 351, 2005
|
|
1J8S
 
 | | PAPG ADHESIN RECEPTOR BINDING DOMAIN-UNBOUND FORM | | Descriptor: | PYELONEPHRITIC ADHESIN | | Authors: | Dodson, K.W, Pinkner, J.S, Rose, T, Magnusson, G, Hultgren, S.J, Waksman, G. | | Deposit date: | 2001-05-22 | | Release date: | 2001-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor.
Cell(Cambridge,Mass.), 105, 2001
|
|
