2B2D
 
 | | RNA stemloop operator from bacteriophage QBETA complexed with an N87S,E89K mutant MS2 capsid | | Descriptor: | 5'-R(*AP*UP*GP*CP*AP*UP*GP*UP*CP*UP*AP*AP*GP*AP*CP*AP*GP*CP*AP*U)-3', Coat protein | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-09-19 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of RNA binding discrimination between bacteriophages Qbeta and MS2
Structure, 14, 2006
|
|
2B2G
 
 | | MS2 Wild-type RNA stemloop complexed with an N87S mutant MS2 capsid | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP*UP*AP*CP*CP*CP*AP*UP*GP*U)-3', Coat protein | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-09-19 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural basis of RNA binding discrimination between bacteriophages Qbeta and MS2
Structure, 14, 2006
|
|
1YP7
 
 | | Van der Waals Interactions Dominate Hydrophobic Association in a Protein Binding Site Occluded From Solvent Water | | Descriptor: | CADMIUM ION, MAJOR URINARY PROTEIN 1 | | Authors: | Barratt, E, Bingham, R.J, Warner, D.J, Laughton, C.A, Phillips, S.E.V, Homans, S.W. | | Deposit date: | 2005-01-30 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Van der Waals Interactions Dominate Ligand-Protein Association in a Protein Binding Site Occluded from Solvent Water
J.Am.Chem.Soc., 127, 2005
|
|
2B2E
 
 | | RNA stemloop from bacteriophage MS2 complexed with an N87S,E89K mutant MS2 capsid | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP*UP*AP*CP*CP*CP*AP*UP*GP*U)-3', Coat protein | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-09-19 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of RNA binding discrimination between bacteriophages Qbeta and MS2
Structure, 14, 2006
|
|
1ZSE
 
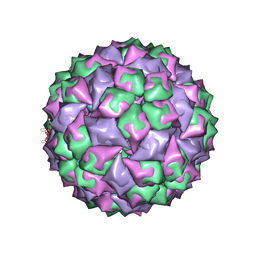 | | RNA stemloop from bacteriophage Qbeta complexed with an N87S mutant MS2 Capsid | | Descriptor: | Coat protein, RNA HAIRPIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-05-24 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2
Structure, 14, 2006
|
|
4PGM
 
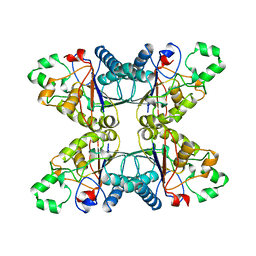 | |
3DC5
 
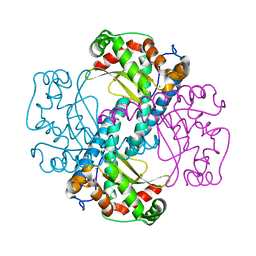 | | Crystal Structure of a manganese superoxide dismutases from Caenorhabditis elegans | | Descriptor: | MALONATE ION, MANGANESE (II) ION, Superoxide dismutase [Mn] 2 | | Authors: | Trinh, C.H, Hunter, T, Stewart, E.E, Phillips, S.E.V, Hunter, G.J. | | Deposit date: | 2008-06-03 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Purification, crystallization and X-ray structures of the two manganese superoxide dismutases from Caenorhabditis elegans
Acta Crystallogr.,Sect.F, 64, 2008
|
|
3DC6
 
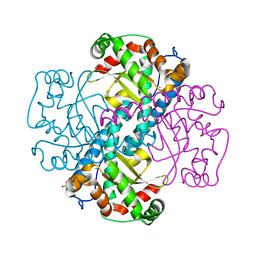 | | Crystal Structure of a manganese superoxide dismutases from Caenorhabditis elegans | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Superoxide dismutase [Mn] 1 | | Authors: | Trinh, C.H, Hunter, T, Stewart, E.E, Phillips, S.E.V, Hunter, G.J. | | Deposit date: | 2008-06-03 | | Release date: | 2009-06-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Purification, crystallization and X-ray structures of the two manganese superoxide dismutases from Caenorhabditis elegans
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1BQ3
 
 | |
4CWE
 
 | |
4CWC
 
 | |
4CIJ
 
 | |
5A4I
 
 | | The mechanism of Hydrogen activation by NiFE-hydrogenases | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Evans, R.M, Brooke, E.J, Wehlin, S.A.M, Nomerotskaia, E, Sargent, F, Carr, S.C, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2015-06-10 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Mechanism of hydrogen activation by [NiFe] hydrogenases.
Nat. Chem. Biol., 12, 2016
|
|
5ADU
 
 | | The Mechanism of Hydrogen Activation by NiFe-hydrogenases | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Evans, R, Brooke, E.J, Wehlin, S.A, Nomerotskaia, E, Sargent, F, Carr, S.B, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2015-08-24 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mechanism of hydrogen activation by [NiFe] hydrogenases.
Nat. Chem. Biol., 12, 2016
|
|
5A4F
 
 | | The mechanism of Hydrogen Activation by NiFe-hydrogenases. | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Evans, R.M, Brooke, E.J, Wehlin, S.A.M, Nomerotskaia, E, Sargent, F, Carr, S.B, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2015-06-09 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mechanism of hydrogen activation by [NiFe] hydrogenases.
Nat. Chem. Biol., 12, 2016
|
|
2P5L
 
 | | Crystal structure of a dimer of N-terminal domains of AhrC in complex with an 18bp DNA operator site | | Descriptor: | Arginine repressor, DNA (5'-D(*DCP*DAP*DTP*DGP*DAP*DAP*DTP*DAP*DAP*DAP*DAP*DAP*DTP*DTP*DCP*DAP*DAP*DG)-3'), DNA (5'-D(*DCP*DTP*DTP*DGP*DAP*DAP*DTP*DTP*DTP*DTP*DTP*DAP*DTP*DTP*DCP*DAP*DTP*DG)-3'), ... | | Authors: | Garnett, J.A, Marincs, F, Baumberg, S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 2007-03-15 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and function of the arginine repressor-operator complex from Bacillus subtilis.
J.Mol.Biol., 379, 2008
|
|
2OZQ
 
 | | Crystal Structure of apo-MUP | | Descriptor: | CADMIUM ION, Novel member of the major urinary protein (Mup) gene family, SODIUM ION | | Authors: | Dennis, C.A, Homans, S.W, Phillips, S.E.V, Syme, N.R. | | Deposit date: | 2007-02-27 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Origin of heat capacity changes in a "nonclassical" hydrophobic interaction.
Chembiochem, 8, 2007
|
|
2P5M
 
 | | C-terminal domain hexamer of AhrC bound with L-arginine | | Descriptor: | ARGININE, Arginine repressor | | Authors: | Garnett, J.A, Baumberg, S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 2007-03-15 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the C-terminal effector-binding domain of AhrC bound to its corepressor L-arginine.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2P5K
 
 | | Crystal structure of the N-terminal domain of AhrC | | Descriptor: | Arginine repressor | | Authors: | Garnett, J.A, Baumberg, S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 2007-03-15 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A high-resolution structure of the DNA-binding domain of AhrC, the arginine repressor/activator protein from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
5MSF
 
 | | MS2 PROTEIN CAPSID/RNA COMPLEX | | Descriptor: | 5'-R(*CP*CP*GP*GP*AP*GP*GP*AP*UP*CP*AP*CP*CP*AP*CP*GP*GP*G)-3', MS2 PROTEIN CAPSID | | Authors: | Rowsell, S, Stonehouse, N.J, Convery, M.A, Adams, C.J, Ellington, A.D, Hirao, I, Peabody, D.S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 1998-05-15 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of a series of RNA aptamers complexed to the same protein target.
Nat.Struct.Biol., 5, 1998
|
|
2W0Q
 
 | | E. coli copper amine oxidase in complex with Xenon | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE, ... | | Authors: | Pirrat, P, Smith, M.A, Pearson, A.R, McPherson, M.J, Phillips, S.E.V. | | Deposit date: | 2008-08-20 | | Release date: | 2008-12-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of a Xenon Derivative of Escherichia Coli Copper Amine Oxidase: Confirmation of the Proposed Oxygen-Entry Pathway.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2VZ1
 
 | | Premat-galactose oxidase | | Descriptor: | ACETATE ION, CALCIUM ION, GALACTOSE OXIDASE | | Authors: | Rogers, M.S, Hurtado-Guerrero, R, Firbank, S.J, Halcrow, M.A, Dooley, D.M, Phillips, S.E.V, Knowles, P.F, McPherson, M.J. | | Deposit date: | 2008-07-29 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Cross-Link Formation of the Cysteine 228-Tyrosine 272 Catalytic Cofactor of Galactose Oxidase Does not Require Dioxygen.
Biochemistry, 47, 2008
|
|
2WO5
 
 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in space group P21 crystal form I | | Descriptor: | N-ACETYLNEURAMINATE LYASE | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-21 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
2VZ3
 
 | | bleached galactose oxidase | | Descriptor: | ACETATE ION, COPPER (II) ION, GALACTOSE OXIDASE | | Authors: | Rogers, M.S, Hurtado-Guerrero, R, Firbank, S.J, Halcrow, M.A, Dooley, D.M, Phillips, S.E.V, Knowles, P.F, McPherson, M.J. | | Deposit date: | 2008-07-29 | | Release date: | 2008-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Link Formation of the Cysteine 228-Tyrosine 272 Catalytic Cofactor of Galactose Oxidase Does not Require Dioxygen.
Biochemistry, 47, 2008
|
|
2WNN
 
 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in complex with pyruvate in space group P21 | | Descriptor: | N-ACETYLNEURAMINATE LYASE, PENTAETHYLENE GLYCOL, SODIUM ION | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-13 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
