8RAT
 
 | |
7UGR
 
 | | Crystal structure of hyperfolder YFP | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Hyperfolder yellow fluorescent protein, ... | | Authors: | Campbell, B.C, Liu, C.F, Petsko, G.A. | | Deposit date: | 2022-03-25 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Chemically stable fluorescent proteins for advanced microscopy.
Nat.Methods, 19, 2022
|
|
5I91
 
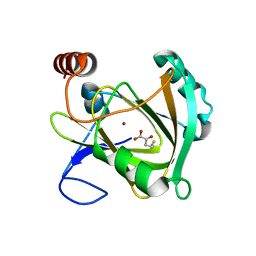 | | Structure of Mouse Acirecutone dioxygenase with to Ni2+ and 2-keto-4-(methylthio)-butyric acid in the active site | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, 4-(METHYLSULFANYL)-2-OXOBUTANOIC ACID, NICKEL (II) ION | | Authors: | Deshpande, A.R, Robinson, H, Wagenpfeil, K, Pochapsky, T.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2016-02-19 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Metal-Dependent Function of a Mammalian Acireductone Dioxygenase.
Biochemistry, 55, 2016
|
|
7RAT
 
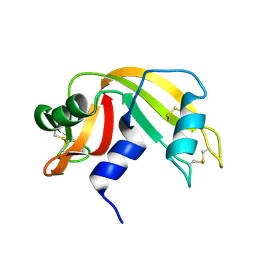 | |
5I8S
 
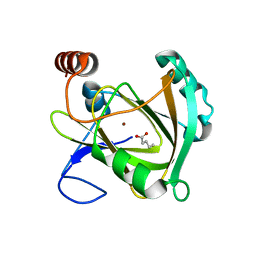 | | Structure of Mouse Acireductone dioxygenase with Ni2+ ion and pentanoic acid in the active site | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, NICKEL (II) ION, PENTANOIC ACID | | Authors: | Deshpande, A.R, Wagenpfeil, K, Pochapsky, T.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2016-02-19 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Metal-Dependent Function of a Mammalian Acireductone Dioxygenase.
Biochemistry, 55, 2016
|
|
7UGS
 
 | |
7UGT
 
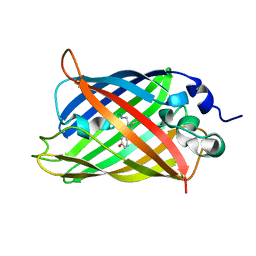 | |
4DM9
 
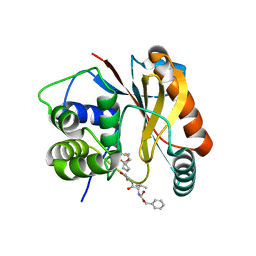 | | The Crystal Structure of Ubiquitin Carboxy-terminal hydrolase L1 (UCHL1) bound to a tripeptide fluoromethyl ketone Z-VAE(OMe)-FMK | | Descriptor: | Tripeptide fluoromethyl ketone inhibitor Z-VAE(OMe)-FMK, Ubiquitin carboxyl-terminal hydrolase isozyme L1 | | Authors: | Davies, C.W, Chaney, J, Korbel, G, Ringe, D, Petsko, G.A, Ploegh, H, Das, C. | | Deposit date: | 2012-02-07 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The co-crystal structure of ubiquitin carboxy-terminal hydrolase L1 (UCHL1) with a tripeptide fluoromethyl ketone (Z-VAE(OMe)-FMK).
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6XNO
 
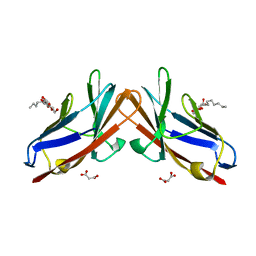 | | Crystal structure of E99A mutant of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, MALONIC ACID, octyl beta-D-glucopyranoside | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Bonsor, D, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2020-07-03 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Commun Biol, 4, 2021
|
|
6RHN
 
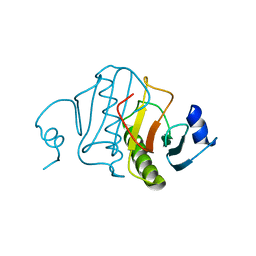 | | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN (HINT) FROM RABBIT WITHOUT NUCLEOTIDE | | Descriptor: | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN | | Authors: | Brenner, C, Garrison, P, Gilmour, J, Peisach, D, Ringe, D, Petsko, G.A, Lowenstein, J.M. | | Deposit date: | 1997-02-27 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins.
Nat.Struct.Biol., 4, 1997
|
|
6RAT
 
 | |
2YPI
 
 | |
6XNT
 
 | | Crystal structure of I91A mutant of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, octyl beta-D-glucopyranoside | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Bonsor, D, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2020-07-04 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Commun Biol, 4, 2021
|
|
6XNW
 
 | | Crystal structure of V39A mutant of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, NICKEL (II) ION | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Bonsor, D, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2020-07-04 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Commun Biol, 4, 2021
|
|
4GCH
 
 | |
6XO1
 
 | | Crystal structure of N97A mutant of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, MALONIC ACID | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Bonsor, D, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2020-07-05 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.758 Å) | | Cite: | Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Commun Biol, 4, 2021
|
|
5BKM
 
 | | Crystal Structure of Hip1 (Rv2224c) mutant - S228DHA (dehydroalanine) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, Goldfarb, N.E, Doran, M.H, Baikovitz, J, Liu, D, Sun, S, White, A, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2021-03-20 | | Release date: | 2022-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal Structure of Hip1 (Rv2224c) mutant - S228DHA (dehydroalanine)
To Be Published
|
|
1GL3
 
 | | ASPARTATE BETA-SEMIALDEHYDE DEHYDROGENASE IN COMPLEX WITH NADP AND SUBSTRATE ANALOGUE S-METHYL CYSTEINE SULFOXIDE | | Descriptor: | ASPARTATE-SEMIALDEHYDE DEHYDROGENASE, CYSTEINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hadfield, A.T, Kryger, G, Ouyang, J, Ringe, D, Petsko, G.A, Viola, R.E. | | Deposit date: | 2001-08-23 | | Release date: | 2001-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Active Site Analysis of the Potential Antimicrobial Target Aspartate Semialdehyde Dehydrogenase.
Biochemistry, 40, 2001
|
|
2NSX
 
 | | Structure of acid-beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, GLYCEROL, ... | | Authors: | Lieberman, R.L, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-11-06 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of acid beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease.
Nat.Chem.Biol., 3, 2007
|
|
2NT1
 
 | | Structure of acid-beta-glucosidase at neutral pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, PHOSPHATE ION | | Authors: | Lieberman, R.L, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-11-06 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of acid beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease.
Nat.Chem.Biol., 3, 2007
|
|
2NT0
 
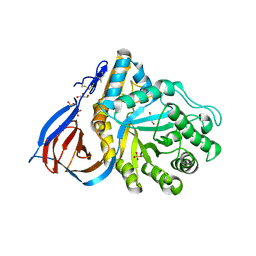 | | Acid-beta-glucosidase low pH, glycerol bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Glucosylceramidase, ... | | Authors: | Lieberman, R.L, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-11-06 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of acid beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease.
Nat.Chem.Biol., 3, 2007
|
|
3FFS
 
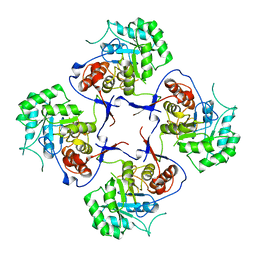 | | The Crystal Structure of Cryptosporidium parvum Inosine-5'-Monophosphate Dehydrogenase | | Descriptor: | Inosine-5-monophosphate dehydrogenase | | Authors: | Riera, T.V, D'Aquino, J.A, Lu, J, Petsko, G.A, Hedstrom, L. | | Deposit date: | 2008-12-04 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The structural basis of Cryptosporidium -specific IMP dehydrogenase inhibitor selectivity
J.Am.Chem.Soc., 132, 2010
|
|
5EAA
 
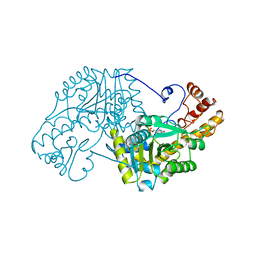 | | ASPARTATE AMINOTRANSFERASE FROM E. COLI, C191S MUTATION | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jeffery, C.J, Gloss, L.M, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-12-29 | | Release date: | 2000-10-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The role of residues outside the active site: structural basis for function of C191 mutants of Escherichia coli aspartate aminotransferase.
Protein Eng., 13, 2000
|
|
5DZL
 
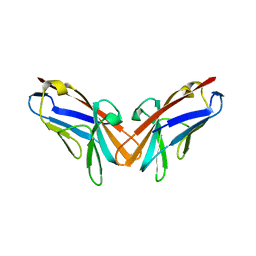 | | Crystal structure of the protein human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1 | | Authors: | Huang, Y.H, Russell, A, Gandhi, A.K, Kondo, Y, Chen, Q, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2015-09-25 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4006 Å) | | Cite: | CEACAM1 regulates TIM-3-mediated tolerance and exhaustion.
Nature, 517, 2015
|
|
3VGN
 
 | | Crystal Structure of Ketosteroid Isomerase D40N from Pseudomonas putida (pKSI) with bound 3-fluoro-4-nitrophenol | | Descriptor: | 3-fluoro-4-nitrophenol, Steroid Delta-isomerase | | Authors: | Caaveiro, J.M.M, Pybus, B, Ringe, D, Petsko, G.A, Sigala, P.A. | | Deposit date: | 2011-08-16 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Quantitative dissection of hydrogen bond-mediated proton transfer in the ketosteroid isomerase active site
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
