1YSY
 
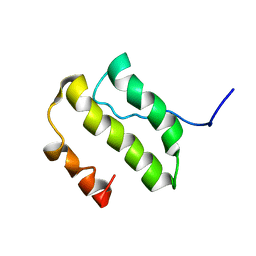 | | NMR Structure of the nonstructural Protein 7 (nsP7) from the SARS CoronaVirus | | Descriptor: | Replicase polyprotein 1ab (pp1ab) (ORF1AB) | | Authors: | Peti, W, Herrmann, T, Johnson, M.A, Kuhn, P, Stevens, R.C, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2005-02-09 | | Release date: | 2005-12-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein nsP7.
J.Virol., 79, 2005
|
|
1R73
 
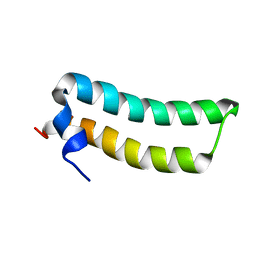 | | Solution Structure of TM1492, the L29 ribosomal protein from Thermotoga maritima | | Descriptor: | 50S ribosomal protein L29 | | Authors: | Peti, W, Etezady-Esfarjani, T, Herrmann, T, Klock, H.E, Lesley, S.A, Wuethrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-10-17 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR for structural proteomics of Thermotoga maritima: Screening and structure determination
J.STRUCT.FUNCT.GENOM., 5, 2004
|
|
1RHX
 
 | |
8U5G
 
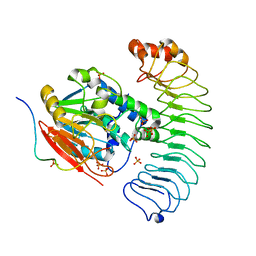 | | Crystal structure of the co-expressed SDS22:PP1:I3 complex | | Descriptor: | E3 ubiquitin-protein ligase PPP1R11, FE (III) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2023-09-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The SDS22:PP1:I3 complex: SDS22 binding to PP1 loosens the active site metal to prime metal exchange.
J.Biol.Chem., 300, 2023
|
|
2AFE
 
 | | Solution Structure of Asl1650, an Acyl Carrier Protein from Anabaena sp. PCC 7120 with a Variant Phosphopantetheinylation-Site Sequence | | Descriptor: | protein Asl1650 | | Authors: | Johnson, M.A, Peti, W, Herrmann, T, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2005-07-25 | | Release date: | 2005-08-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Asl1650, an acyl carrier protein from Anabaena sp. PCC 7120 with a variant phosphopantetheinylation-site sequence
Protein Sci., 15, 2006
|
|
2AFD
 
 | | Solution Structure of Asl1650, an Acyl Carrier Protein from Anabaena sp. PCC 7120 with a Variant Phosphopantetheinylation-Site Sequence | | Descriptor: | protein Asl1650 | | Authors: | Johnson, M.A, Peti, W, Herrmann, T, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2005-07-25 | | Release date: | 2005-08-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Asl1650, an acyl carrier protein from Anabaena sp. PCC 7120 with a variant phosphopantetheinylation-site sequence
Protein Sci., 15, 2006
|
|
4MOV
 
 | | 1.45 A Resolution Crystal Structure of Protein Phosphatase 1 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4503 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MOY
 
 | | Structure of a second nuclear PP1 Holoenzyme, crystal form 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Choy, M.S, Hieke, M, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1953 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MP0
 
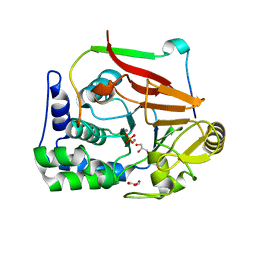 | | Structure of a second nuclear PP1 Holoenzyme, crystal form 2 | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Hieke, M, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1003 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1WCJ
 
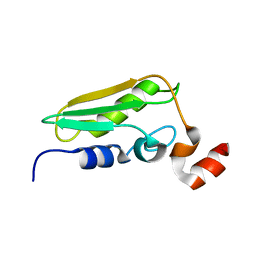 | | Conserved Hypothetical Protein TM0487 from Thermotoga maritima | | Descriptor: | HYPOTHETICAL PROTEIN TM0487 | | Authors: | Almeida, M.S, Peti, W, Herrmann, T, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-11-17 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Conserved Hypothetical Protein Tm0487 from Thermotoga Maritima: Implications for 216 Homologous Duf59 Proteins.
Protein Sci., 14, 2005
|
|
7T0Y
 
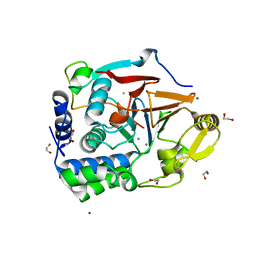 | | The Ribosomal RNA Processing 1B Protein Phosphatase-1 Holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, FLUORIDE ION, ... | | Authors: | Srivastava, G, Page, R, Peti, W. | | Deposit date: | 2021-11-30 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The ribosomal RNA processing 1B:protein phosphatase 1 holoenzyme reveals non-canonical PP1 interaction motifs.
Cell Rep, 41, 2022
|
|
1OJG
 
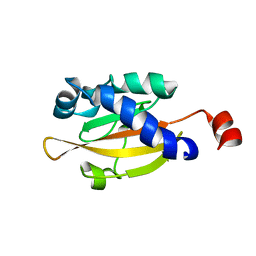 | | Sensory domain of the membraneous two-component fumarate sensor DcuS of E. coli | | Descriptor: | SENSOR PROTEIN DCUS | | Authors: | Pappalardo, L, Janausch, I.G, Vijayan, V, Zientz, E, Junker, J, Peti, W, Zweckstetter, M, Unden, G, Griesinger, C. | | Deposit date: | 2003-07-10 | | Release date: | 2003-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the sensory domain of the membranous two-component fumarate sensor (histidine protein kinase) DcuS of Escherichia coli.
J. Biol. Chem., 278, 2003
|
|
6HKW
 
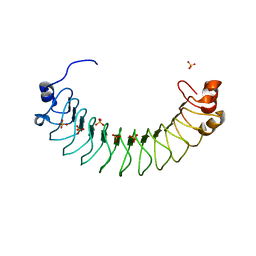 | | Crystal structure of human SDS22 | | Descriptor: | Protein phosphatase 1 regulatory subunit 7, SULFATE ION | | Authors: | Heroes, E, Choy, M.S, Page, R, Peti, W, Ulens, C, Van Meervelt, L, Nys, M, Bollen, M. | | Deposit date: | 2018-09-09 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Guided Exploration of SDS22 Interactions with Protein Phosphatase PP1 and the Splicing Factor BCLAF1.
Structure, 27, 2019
|
|
6UUQ
 
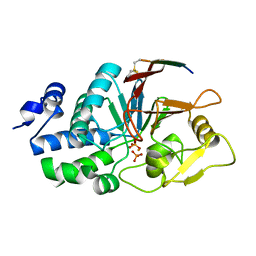 | | Structure of Calcineurin bound to RCAN1 | | Descriptor: | Calcipressin-1, FE (III) ION, PHOSPHATE ION, ... | | Authors: | Sheftic, S, Page, R, Peti, W. | | Deposit date: | 2019-10-31 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | The structure of the RCAN1:CN complex explains the inhibition of and substrate recruitment by calcineurin.
Sci Adv, 6, 2020
|
|
6VRO
 
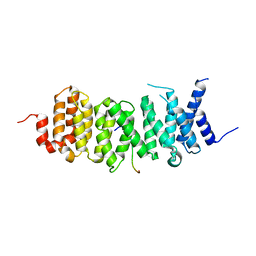 | | The structure of the PP2A B56 subunit AIM1 complex | | Descriptor: | Beta/gamma crystallin domain-containing protein 1, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform | | Authors: | Wang, X, Page, R, Peti, W. | | Deposit date: | 2020-02-08 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A dynamic charge-charge interaction modulates PP2A:B56 substrate recruitment.
Elife, 9, 2020
|
|
5W1E
 
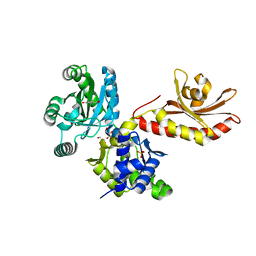 | | PobR in complex with PHB | | Descriptor: | GLYCEROL, P-HYDROXYBENZOIC ACID, Putative transcriptional regulator, ... | | Authors: | Page, R, Peti, W, Lord, D.M, Bajaj, R, Zhang, R. | | Deposit date: | 2017-06-02 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A peculiar IclR family transcription factor regulates para-hydroxybenzoate catabolism in Streptomyces coelicolor.
Nucleic Acids Res., 46, 2018
|
|
5INB
 
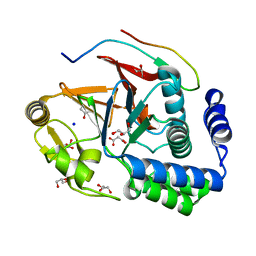 | | RepoMan-PP1g (protein phosphatase 1, gamma isoform) holoenzyme complex | | Descriptor: | Cell division cycle-associated protein 2, GLYCEROL, MALONATE ION, ... | | Authors: | Kumar, G.S, Peti, W, Page, R. | | Deposit date: | 2016-03-07 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Ki-67 and RepoMan mitotic phosphatases assemble via an identical, yet novel mechanism.
Elife, 5, 2016
|
|
3V4Y
 
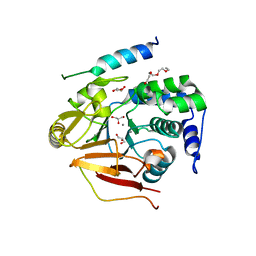 | | Crystal Structure of the first Nuclear PP1 holoenzyme | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Nuclear inhibitor of protein phosphatase 1, ... | | Authors: | Page, R, Peti, W, O'Connell, N.E, Nichols, S. | | Deposit date: | 2011-12-15 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | The Molecular Basis for Substrate Specificity of the Nuclear NIPP1:PP1 Holoenzyme.
Structure, 20, 2012
|
|
5IOH
 
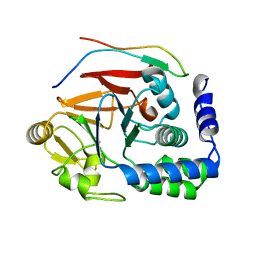 | | RepoMan-PP1a (protein phosphatase 1, alpha isoform) holoenzyme complex | | Descriptor: | Cell division cycle-associated protein 2, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Kumar, G.S, Peti, W, Page, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.566 Å) | | Cite: | The Ki-67 and RepoMan mitotic phosphatases assemble via an identical, yet novel mechanism.
Elife, 5, 2016
|
|
4XPN
 
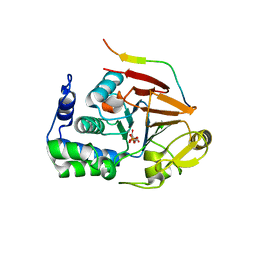 | | Crystal Structure of Protein Phosphate 1 complexed with PP1 binding domain of GADD34 | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2015-01-17 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Structural and Functional Analysis of the GADD34:PP1 eIF2 alpha Phosphatase.
Cell Rep, 11, 2015
|
|
4Y14
 
 | | Structure of protein tyrosine phosphatase 1B complexed with inhibitor (PTP1B:CPT157633) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-bromo-4-[difluoro(phosphono)methyl]-N-methyl-Nalpha-(methylsulfonyl)-L-phenylalaninamide, CHLORIDE ION, ... | | Authors: | Choy, M.S, Connors, C, Page, R, Peti, W. | | Deposit date: | 2015-02-06 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | PTP1B inhibition suggests a therapeutic strategy for Rett syndrome.
J.Clin.Invest., 125, 2015
|
|
6ATD
 
 | |
6ALZ
 
 | | Crystal structure of Protein Phosphatase 1 bound to the natural inhibitor Tautomycetin | | Descriptor: | (2Z)-2-[(1R)-3-{[(2R,3S,4R,7S,8S,11S,13R,16E)-17-ethyl-4,8-dihydroxy-3,7,11,13-tetramethyl-6,15-dioxononadeca-16,18-dien-2-yl]oxy}-1-hydroxy-3-oxopropyl]-3-methylbut-2-enedioic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2017-08-08 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | PP1:Tautomycetin Complex Reveals a Path toward the Development of PP1-Specific Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
3O9X
 
 | | Structure of the E. coli antitoxin MqsA (YgiT/b3021) in complex with its gene promoter | | Descriptor: | DNA (26-MER), GLYCEROL, Uncharacterized HTH-type transcriptional regulator ygiT, ... | | Authors: | Brown, B.L, Peti, W, Page, R. | | Deposit date: | 2010-08-04 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.0999 Å) | | Cite: | Structure of the Escherichia coli Antitoxin MqsA (YgiT/b3021) Bound to Its Gene Promoter Reveals Extensive Domain Rearrangements and the Specificity of Transcriptional Regulation.
J.Biol.Chem., 286, 2011
|
|
1UWD
 
 | | NMR STRUCTURE OF A PROTEIN WITH UNKNOWN FUNCTION FROM THERMOTOGA MARITIMA (TM0487), WHICH BELONGS TO THE DUF59 FAMILY. | | Descriptor: | HYPOTHETICAL PROTEIN TM0487 | | Authors: | Almeida, M.S, Peti, W, Herrmann, T, Wuthrich, K. | | Deposit date: | 2004-02-03 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Conserved Hypothetical Protein Tm0487 from Thermotoga Maritima: Implications for 216 Homologous Duf59 Proteins.
Protein Sci., 14, 2005
|
|
