5FSR
 
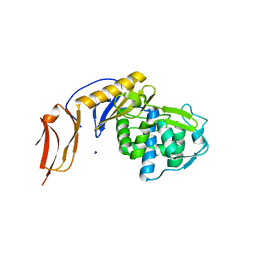 | | Crystal structure of penicillin binding protein 6B from Escherichia coli | | Descriptor: | D-ALANYL-D-ALANINE CARBOXYPEPTIDASE DACD | | Authors: | Peters, K, Kannan, S, Rao, V.A, Bilboy, J, Vollmer, D, Erickson, S.W, Lewis, R.J, Young, K.D, Vollmer, W. | | Deposit date: | 2016-01-07 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Redundancy of Peptidoglycan Carboxypeptidases Ensures Robust Cell Shape Maintenance in Escherichia Coli
Mbio, 7, 2016
|
|
4EO4
 
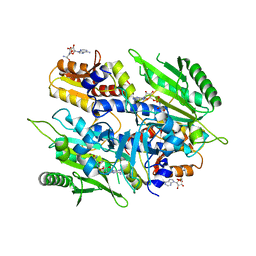 | | Crystal structure of the yeast mitochondrial threonyl-tRNA synthetase (MST1) in complex with seryl sulfamoyl adenylate | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, Threonine--tRNA ligase, mitochondrial, ... | | Authors: | Peterson, K.M, Ling, J, Simonovic, I, Soll, D, Simonovic, M. | | Deposit date: | 2012-04-13 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | The mechanism of pre-transfer editing in yeast mitochondrial threonyl-tRNA synthetase.
J.Biol.Chem., 287, 2012
|
|
3UGQ
 
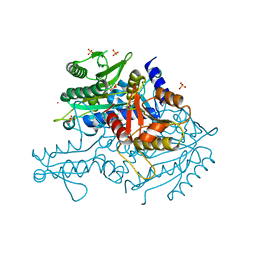 | | Crystal structure of the apo form of the yeast mitochondrial threonyl-tRNA synthetase determined at 2.1 Angstrom resolution | | Descriptor: | POTASSIUM ION, SULFATE ION, Threonyl-tRNA synthetase, ... | | Authors: | Peterson, K.M, Ling, J, Simonovic, I, Cho, C, Soll, D, Simonovic, M. | | Deposit date: | 2011-11-02 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Yeast mitochondrial threonyl-tRNA synthetase recognizes tRNA isoacceptors by distinct mechanisms and promotes CUN codon reassignment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UGT
 
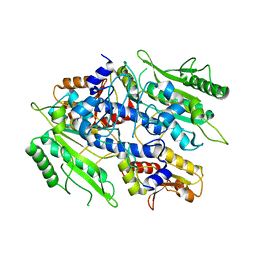 | | Crystal structure of the yeast mitochondrial threonyl-tRNA synthetase - orthorhombic crystal form | | Descriptor: | Threonyl-tRNA synthetase, mitochondrial, ZINC ION | | Authors: | Peterson, K.M, Ling, J, Simonovic, I, Cho, C, Soll, D, Simonovic, M. | | Deposit date: | 2011-11-02 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Yeast mitochondrial threonyl-tRNA synthetase recognizes tRNA isoacceptors by distinct mechanisms and promotes CUN codon reassignment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1HXY
 
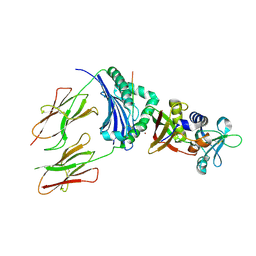 | | CRYSTAL STRUCTURE OF STAPHYLOCOCCAL ENTEROTOXIN H IN COMPLEX WITH HUMAN MHC CLASS II | | Descriptor: | ENTEROTOXIN H, HEMAGGLUTININ, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Petersson, K, Hakansson, M, Nilsson, H, Forsberg, G, Svensson, L.A, Liljas, A, Walse, B. | | Deposit date: | 2001-01-17 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Superantigen Bound to MHC Class II Displays Zinc and Peptide Dependence
Embo J., 20, 2001
|
|
1LO5
 
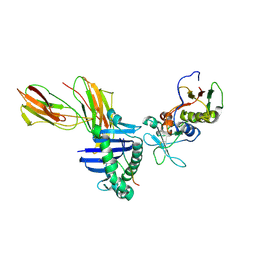 | | Crystal structure of the D227A variant of Staphylococcal enterotoxin A in complex with human MHC class II | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DR-1 beta chain, ... | | Authors: | Petersson, K, Thunnissen, M, Forsberg, G, Walse, B. | | Deposit date: | 2002-05-06 | | Release date: | 2002-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of a SEA Variant in Complex with MHC Class II Reveals the Ability of SEA to Crosslink MHC Molecules
Structure, 10, 2002
|
|
3UH0
 
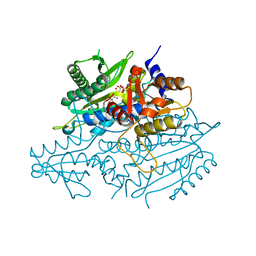 | | Crystal structure of the yeast mitochondrial threonyl-tRNA synthetase (MST1) in complex with threonyl sulfamoyl adenylate | | Descriptor: | 5'-O-(N-(L-THREONYL)-SULFAMOYL)ADENOSINE, SULFATE ION, Threonyl-tRNA synthetase, ... | | Authors: | Peterson, K.M, Ling, J, Simonovic, I, Cho, C, Soll, D, Simonovic, M. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Yeast mitochondrial threonyl-tRNA synthetase recognizes tRNA isoacceptors by distinct mechanisms and promotes CUN codon reassignment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6ZTG
 
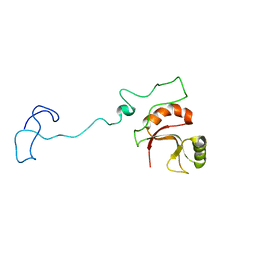 | | Spor protein DedD | | Descriptor: | Cell division protein DedD | | Authors: | Pazos, M, Peters, K, Boes, A, Safaei, Y, Kenward, C, Caveney, N.A, Laguri, C, Breukink, E, Strynadka, N.C.J, Simorre, J.P, Terrak, M, Vollmer, W. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | SPOR Proteins Are Required for Functionality of Class A Penicillin-Binding Proteins in Escherichia coli.
Mbio, 11, 2020
|
|
1PJ8
 
 | | Structure of a ternary complex of proteinase K, mercury and a substrate-analogue hexapeptide at 2.2 A resolution | | Descriptor: | 6-residue peptide (N-Ac-PAPFPA-NH2), MERCURY (II) ION, Proteinase K | | Authors: | Saxena, A.K, Singh, T.P, Peters, K, Fittkau, S, Visanji, M, Wilson, K.S, Betzel, C. | | Deposit date: | 2003-06-02 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a ternary complex of proteinase K, mercury, and a substrate-analogue hexa-peptide at 2.2 A resolution
Proteins, 25, 1996
|
|
1PFG
 
 | | Strategy to design inhibitors: Structure of a complex of Proteinase K with a designed octapeptide inhibitor N-Ac-Pro-Ala-Pro-Phe-DAla-Ala-Ala-Ala-NH2 at 2.5A resolution | | Descriptor: | N-Ac-PAPFAAAA-NH2, Proteinase K | | Authors: | Saxena, A.K, Singh, T.P, Peters, K, Fittkau, S, Betzel, C. | | Deposit date: | 2003-05-27 | | Release date: | 2003-06-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Strategy to design peptide inhibitors: structure of a complex of proteinase K with a designed octapeptide inhibitor N-Ac-Pro-Ala-Pro-Phe-DAla-Ala-Ala-Ala-NH2 at 2.5 A resolution.
Protein Sci., 5, 1996
|
|
1PEK
 
 | | STRUCTURE OF THE COMPLEX OF PROTEINASE K WITH A SUBSTRATE-ANALOGUE HEXA-PEPTIDE INHIBITOR AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | D-DAL-ALA-NH2, PEPTIDE PRO-ALA-PRO-PHE, PROTEINASE K | | Authors: | Betzel, C, Singh, T.P, Visanji, M, Peters, K, Fittkau, S, Saenger, W, Wilson, K.S. | | Deposit date: | 1993-01-19 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the complex of proteinase K with a substrate analogue hexapeptide inhibitor at 2.2-A resolution.
J.Biol.Chem., 268, 1993
|
|
6FQB
 
 | | MurT/GatD peptidoglycan amidotransferase complex from Streptococcus pneumoniae R6 | | Descriptor: | Cobyric acid synthase, GLUTAMINE, Mur ligase family protein | | Authors: | Morlot, C, Contreras-Martel, C, Leisico, F, Straume, D, Peters, K, Hegnar, O.A, Simon, N, Villard, A.M, Breukink, E, Gravier-Pelletier, C, Le Corre, L, Vollmer, W, Pietrancosta, N, Havarstein, L.S, Zapun, A. | | Deposit date: | 2018-02-13 | | Release date: | 2018-08-22 | | Last modified: | 2018-11-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the essential peptidoglycan amidotransferase MurT/GatD complex from Streptococcus pneumoniae.
Nat Commun, 9, 2018
|
|
2H02
 
 | | Structural studies of protein tyrosine phosphatase beta catalytic domain in complex with inhibitors | | Descriptor: | Protein tyrosine phosphatase, receptor type, B,, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M, Gray, J.L, Peters, K.G, Maier, M.B, Amarasinghe, K.D, Clark, C.M, Nichols, R. | | Deposit date: | 2006-05-13 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and synthesis of potent, non-peptidic inhibitors of HPTPbeta.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2H04
 
 | | Structural studies of protein tyrosine phosphatase beta catalytic domain in complex with inhibitors | | Descriptor: | Protein tyrosine phosphatase, receptor type, B,, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M, Gray, J.L, Peters, K.G, Maier, M.B, Amarasinghe, K.D, Clark, C.M, Nichols, R. | | Deposit date: | 2006-05-13 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and synthesis of potent, non-peptidic inhibitors of HPTPbeta.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2H03
 
 | | Structural studies of protein tyrosine phosphatase beta catalytic domain in complex with inhibitors | | Descriptor: | (4-{4-[(TERT-BUTOXYCARBONYL)AMINO]-2,2-BIS(ETHOXYCARBONYL)BUTYL}PHENYL)SULFAMIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M, Gray, J.L, Peters, K.G, Maier, M.B, Amarasinghe, K.D, Clark, C.M, Nichols, R. | | Deposit date: | 2006-05-13 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design and synthesis of potent, non-peptidic inhibitors of HPTPbeta.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4UDU
 
 | | Crystal structure of staphylococcal enterotoxin E in complex with a T cell receptor | | Descriptor: | ENTEROTOXIN TYPE E, PROTEIN TRBV7-9, T-CELL RECEPTOR BETA-2 CHAIN C REGION, ... | | Authors: | Rodstrom, K.E.J, Regenthal, P, Lindkvist-Petersson, K. | | Deposit date: | 2014-12-11 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Staphylococcal Enterotoxin E in Complex with Tcr Defines the Role of Tcr Loop Positioning in Superantigen Recognition.
Plos One, 10, 2015
|
|
4UDT
 
 | | T cell receptor (TRAV22,TRBV7-9) structure | | Descriptor: | GLYCEROL, PROTEIN TRBV7-9, T-CELL RECEPTOR BETA-2 CHAIN C REGION, ... | | Authors: | Rodstrom, K.E.J, Regenthal, P, Lindkvist-Petersson, K. | | Deposit date: | 2014-12-11 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of Staphylococcal Enterotoxin E in Complex with Tcr Defines the Role of Tcr Loop Positioning in Superantigen Recognition.
Plos One, 10, 2015
|
|
6EPM
 
 | | Ras guanine nucleotide exchange factor SOS1 (Rem-cdc25) in complex with KRAS(G12C) and fragment screening hit F1 | | Descriptor: | (1-phenyl-5,6-dihydro-4~{H}-cyclopenta[c]pyrazol-3-yl)methanamine, GLYCEROL, GTPase KRas, ... | | Authors: | Hillig, R.C, Moosmayer, D, Hilpmann, A, Bader, B, Schroeder, J, Wortmann, L, Sautier, B, Kahmann, J, Wegener, D, Briem, H, Petersen, K, Badock, V. | | Deposit date: | 2017-10-12 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6EPP
 
 | | RAS GUANINE EXCHANGE FACTOR SOS1 (REM-CDC25) IN COMPLEX WITH KRAS(G12C) AND FRAGMENT SCREENING HIT F4 | | Descriptor: | GLYCEROL, GTPase KRas, Son of sevenless homolog 1, ... | | Authors: | Hillig, R.C, Moosmayer, D, Hilpmann, A, Bader, B, Schroeder, J, Wortmann, L, Sautier, B, Kahmann, J, Wegener, D, Briem, H, Petersen, K, Badock, V. | | Deposit date: | 2017-10-12 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5FK9
 
 | |
5FKA
 
 | | Crystal structure of staphylococcal enterotoxin E in complex with a T cell receptor | | Descriptor: | STAPHYLOCOCCAL ENTEROTOXIN E, T CELL RECEPTOR ALPHA CHAIN, T CELL RECEPTOR BETA CHAIN, ... | | Authors: | Rodstrom, K.E.J, Regenthal, P, Lindkvist-Petersson, K. | | Deposit date: | 2015-10-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two Common Structural Motifs for Tcr Recognition by Staphylococcal Enterotoxins.
Sci.Rep., 6, 2016
|
|
8C9H
 
 | | AQP7_inhibitor | | Descriptor: | Aquaporin-7, ethyl 4-[(4-pyrazol-1-ylphenyl)methylcarbamoylamino]benzoate | | Authors: | Huang, P, Venskutonyte, R, Gourdon, P, Lindkvist-Petersson, K. | | Deposit date: | 2023-01-22 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis for human aquaporin inhibition.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7ZQI
 
 | | MHC class I from a wild bird in complex with a nonameric peptide P2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Eltschkner, S, Mellinger, S, Buus, S, Nielsen, M, Paulsson, K.M, Lindkvist-Petersson, K, Westerdahl, H. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of songbird MHC class I reveals antigen binding that is flexible at the N-terminus and static at the C-terminus.
Front Immunol, 14, 2023
|
|
7ZQJ
 
 | | MHC class I from a wild bird in complex with a nonameric peptide P3 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Eltschkner, S, Mellinger, S, Buus, S, Nielsen, M, Paulsson, K.M, Lindkvist-Petersson, K, Westerdahl, H. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of songbird MHC class I reveals antigen binding that is flexible at the N-terminus and static at the C-terminus.
Front Immunol, 14, 2023
|
|
6QZJ
 
 | | Crystal structure of human Aquaporin 7 at 2.2 A resolution | | Descriptor: | Aquaporin-7, GLYCEROL, PHOSPHATE ION | | Authors: | de Mare, S.W.-H, Venskutonyte, R, Eltschkner, S, Lindkvist-Petersson, K. | | Deposit date: | 2019-03-11 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Glycerol Efflux and Selectivity of Human Aquaporin 7.
Structure, 28, 2020
|
|
