6WW9
 
 | | Crystal structure of human REV7(R124A)-SHLD3(35-58) complex | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, Shieldin complex subunit 3 | | Authors: | Xie, W, Patel, D.J. | | Deposit date: | 2020-05-08 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular mechanisms of assembly and TRIP13-mediated remodeling of the human Shieldin complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1SLO
 
 | | FIRST STEM LOOP OF THE SL1 RNA FROM CAENORHABDITIS ELEGANS, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | RNA (5'-R(*UP*UP*AP*CP*CP*CP*AP*AP*GP*UP*UP*UP*GP*AP*GP*GP*UP*AP*A)-3') | | Authors: | Greenbaum, N.L, Radhakrishnan, I, Patel, D.J, Hirsh, D. | | Deposit date: | 1996-05-24 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the donor site of a trans-splicing RNA.
Structure, 4, 1996
|
|
1SLP
 
 | | FIRST STEM LOOP OF THE SL1 RNA FROM CAENORHABDITIS ELEGANS, NMR, 16 STRUCTURES | | Descriptor: | RNA (5'-R(*UP*UP*AP*CP*CP*CP*AP*AP*GP*UP*UP*UP*GP*AP*GP*GP*UP*AP*A)-3') | | Authors: | Greenbaum, N.L, Radhakrishnan, I, Patel, D.J, Hirsh, D. | | Deposit date: | 1996-05-24 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the donor site of a trans-splicing RNA.
Structure, 4, 1996
|
|
6WWA
 
 | | Crystal structure of human SHLD2-SHLD3-REV7 complex | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, Shieldin complex subunit 2,Shieldin complex subunit 3 chimera | | Authors: | Xie, W, Patel, D.J. | | Deposit date: | 2020-05-08 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular mechanisms of assembly and TRIP13-mediated remodeling of the human Shieldin complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3OXE
 
 | | crystal structure of glycine riboswitch, Mn2+ soaked | | Descriptor: | GLYCINE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3OX0
 
 | |
3PT6
 
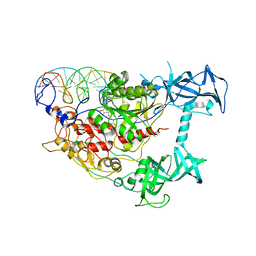 | | Crystal structure of mouse DNMT1(650-1602) in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
3Q0D
 
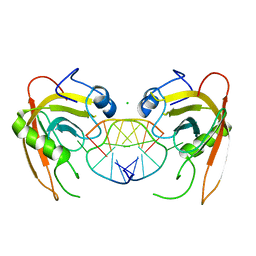 | | Crystal structure of SUVH5 SRA- hemi methylated CG DNA complex | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*TP*GP*AP*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*AP*(5CM)P*GP*TP*CP*AP*G)-3'), ... | | Authors: | Eerappa, R, Simanshu, D.K, Patel, D.J. | | Deposit date: | 2010-12-15 | | Release date: | 2011-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3704 Å) | | Cite: | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
3Q0B
 
 | |
3Q0F
 
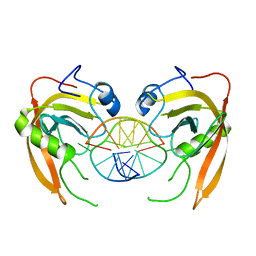 | | Crystal structure of SUVH5 SRA- methylated CHH DNA complex | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*GP*GP*AP*GP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*(5CM)P*CP*TP*CP*AP*G)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Eerappa, R, Simanshu, D.K, Patel, D.J. | | Deposit date: | 2010-12-15 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
3PTA
 
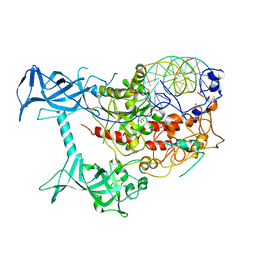 | | Crystal structure of human DNMT1(646-1600) in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
3QZT
 
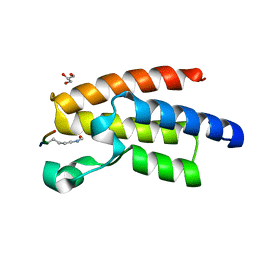 | | Crystal Structure of BPTF bromo in complex with histone H4K16ac - Form II | | Descriptor: | GLYCEROL, Histone H4, Nucleosome-remodeling factor subunit BPTF | | Authors: | Li, H, Ruthenburg, A.J, Patel, D.J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions.
Cell(Cambridge,Mass.), 145, 2011
|
|
3QZS
 
 | |
3RB3
 
 | |
3RBD
 
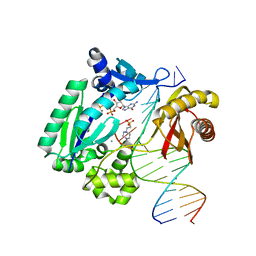 | |
3QZV
 
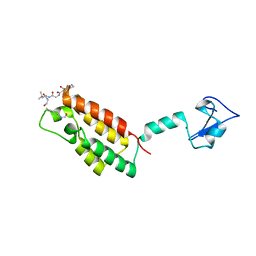 | | Crystal Structure of BPTF PHD-linker-bromo in complex with histone H4K12ac peptide | | Descriptor: | Histone H4, Nucleosome-remodeling factor subunit BPTF, ZINC ION | | Authors: | Li, H, Ruthenburg, A.J, Patel, D.J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions.
Cell(Cambridge,Mass.), 145, 2011
|
|
3O7V
 
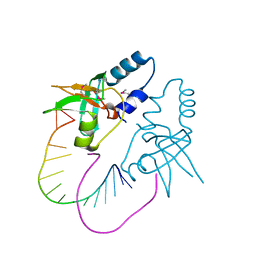 | | Crystal Structure of human Hiwi1 (V361M) PAZ domain (residues 277-399) in complex with 14-mer RNA (12-bp + 2-nt overhang) containing 2'-OCH3 at its 3'-end | | Descriptor: | Piwi-like protein 1, RNA (5'-R(*GP*CP*GP*AP*AP*UP*AP*UP*UP*CP*GP*CP*UP*(OMU))-3') | | Authors: | Tian, Y, Simanshu, D.K, Ma, J.-B, Patel, D.J. | | Deposit date: | 2010-08-01 | | Release date: | 2011-01-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inaugural Article: Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3O6E
 
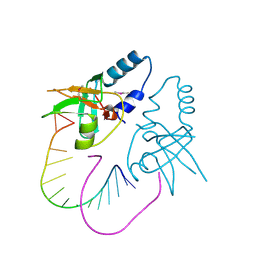 | | Crystal Structure of human Hiwi1 PAZ domain (residues 277-399) in complex with 14-mer RNA (12-bp + 2-nt overhang) containing 2'-OCH3 at its 3'-end | | Descriptor: | Piwi-like protein 1, RNA (5'-R(*GP*CP*GP*AP*AP*UP*AP*UP*UP*CP*GP*CP*UP*(OMU))-3') | | Authors: | Tian, Y, Simanshu, D.K, Ma, J.-B, Patel, D.J. | | Deposit date: | 2010-07-28 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Inaugural Article: Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OWW
 
 | |
6XL1
 
 | | crystal structure of cA4-activated Card1(D294N) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Card1, MANGANESE (II) ION, ... | | Authors: | Rostol, J, Xie, W, Patel, D.J, Marraffini, L. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Card1 nuclease provides defence during type III CRISPR immunity.
Nature, 590, 2021
|
|
3PT9
 
 | | Crystal structure of mouse DNMT1(731-1602) in the free state | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
3RB0
 
 | |
3RB6
 
 | |
3RBE
 
 | |
3RAQ
 
 | |
