2AU0
 
 | | Unmodified preinsertion binary complex | | 分子名称: | 5'-D(*CP*TP*AP*AP*CP*G*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*GP*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*(DDG))-3', CALCIUM ION, ... | | 著者 | Rechkoblit, O, Malinina, L, Cheng, Y, Kuryavyi, V, Broyde, S, Geacintov, N.E, Patel, D.J. | | 登録日 | 2005-08-26 | | 公開日 | 2006-01-10 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Stepwise Translocation of Dpo4 Polymerase during Error-Free Bypass of an oxoG Lesion
Plos Biol., 4, 2006
|
|
2ATL
 
 | | Unmodified Insertion Ternary Complex | | 分子名称: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*T*AP*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*GP*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*(DDG))-3', ... | | 著者 | Rechkoblit, O, Malinina, L, Cheng, Y, Kuryavyi, V, Broyde, S, Geacintov, N.E, Patel, D.J. | | 登録日 | 2005-08-25 | | 公開日 | 2006-01-10 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Stepwise Translocation of Dpo4 Polymerase during Error-Free Bypass of an oxoG Lesion
Plos Biol., 4, 2006
|
|
3D2S
 
 | |
4IUP
 
 | |
4IUQ
 
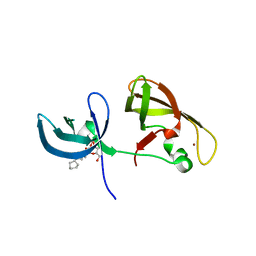 | | crystal structure of SHH1 SAWADEE domain | | 分子名称: | CYMAL-4, SAWADEE HOMEODOMAIN HOMOLOG 1, ZINC ION | | 著者 | Du, J, Patel, D.J. | | 登録日 | 2013-01-21 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.809 Å) | | 主引用文献 | Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1.
Nature, 498, 2013
|
|
2EVS
 
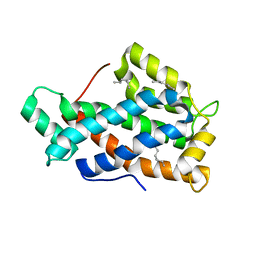 | | Crystal structure of human Glycolipid Transfer Protein complexed with n-hexyl-beta-D-glucoside | | 分子名称: | DECANE, Glycolipid transfer protein, HEXANE, ... | | 著者 | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | 登録日 | 2005-10-31 | | 公開日 | 2006-11-14 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
2JMJ
 
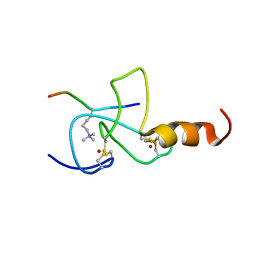 | | NMR solution structure of the PHD domain from the yeast YNG1 protein in complex with H3(1-9)K4me3 peptide | | 分子名称: | Histone H3, Protein YNG1, ZINC ION | | 著者 | Ilin, S, Taverna, S.D, Rogers, R.S, Tanny, J.C, Lavender, H, Li, H, Baker, L, Boyle, J, Blair, L.P, Chait, B.T, Patel, D.J, Aitchison, J.D, Tackett, A.J, Allis, C.D. | | 登録日 | 2006-11-15 | | 公開日 | 2007-07-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs
Mol.Cell, 24, 2006
|
|
3D2Q
 
 | |
4JOL
 
 | |
3D1B
 
 | |
2JMI
 
 | | NMR solution structure of PHD finger fragment of Yeast Yng1 protein in free state | | 分子名称: | Protein YNG1, ZINC ION | | 著者 | Ilin, S, Taverna, S.D, Rogers, R.S, Tanny, J.C, Lavender, H, Li, H, Baker, L, Boyle, J, Blair, L.P, Chait, B.T, Patel, D.J, Aitchison, J.D, Tackett, A.J, Allis, C.D. | | 登録日 | 2006-11-15 | | 公開日 | 2007-07-03 | | 最終更新日 | 2024-10-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs
Mol.Cell, 24, 2006
|
|
4K84
 
 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 16:0 ceramide-1-phosphate (16:0-C1P) | | 分子名称: | (2S,3R,4E)-2-(hexadecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | 著者 | Simanshu, D.K, Brown, R.E, Patel, D.J. | | 登録日 | 2013-04-17 | | 公開日 | 2013-07-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.897 Å) | | 主引用文献 | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4KBS
 
 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 12:0 phosphatidic acid (12:0 PA) | | 分子名称: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1,2-ETHANEDIOL, Glycolipid transfer protein domain-containing protein 1 | | 著者 | Simanshu, D.K, Brown, R.E, Patel, D.J. | | 登録日 | 2013-04-23 | | 公開日 | 2013-07-17 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.898 Å) | | 主引用文献 | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4KBR
 
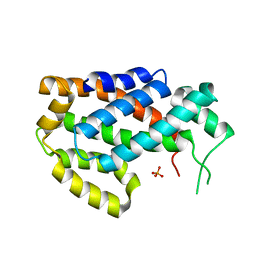 | |
2CNX
 
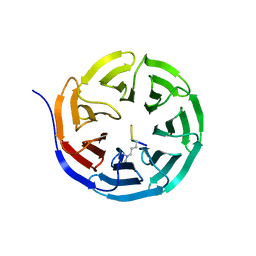 | | WDR5 and Histone H3 Lysine 4 dimethyl complex at 2.1 angstrom | | 分子名称: | HISTONE H3 DIMETHYL-LYSINE 4, WD-REPEAT PROTEIN 5 | | 著者 | Ruthenburg, A.J, Wang, W, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | 登録日 | 2006-05-25 | | 公開日 | 2006-07-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Histone H3 Recognition and Presentation by the Wdr5 Module of the Mll1 Complex
Nat.Struct.Mol.Biol., 13, 2006
|
|
2CO0
 
 | | WDR5 and unmodified Histone H3 complex at 2.25 Angstrom | | 分子名称: | HISTONE H3 DIMETHYL-LYSINE 4, WD-REPEAT PROTEIN 5 | | 著者 | Ruthenburg, A.J, Wang, W, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | 登録日 | 2006-05-25 | | 公開日 | 2006-07-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Histone H3 Recognition and Presentation by the Wdr5 Module of the Mll1 Complex
Nat.Struct.Mol.Biol., 13, 2006
|
|
2ASL
 
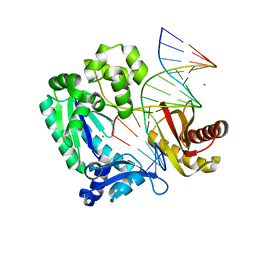 | | oxoG-modified Postinsertion Binary Complex | | 分子名称: | 5'-D(*CP*T*AP*AP*CP*(8OG)P*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*GP*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*(DOC))-3', CALCIUM ION, ... | | 著者 | Rechkoblit, O, Malinina, L, Cheng, Y, Kuryavyi, V, Broyde, S, Geacintov, N.E, Patel, D.J. | | 登録日 | 2005-08-23 | | 公開日 | 2006-01-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Stepwise Translocation of Dpo4 Polymerase during Error-Free Bypass of an oxoG Lesion
Plos Biol., 4, 2006
|
|
4KF6
 
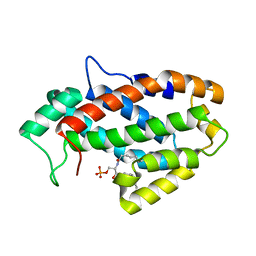 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 8:0 Ceramide-1-Phosphate (8:0-C1P) | | 分子名称: | (2S,3R,4E)-3-hydroxy-2-(octanoylamino)octadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | 著者 | Simanshu, D.K, Brown, R.E, Patel, D.J. | | 登録日 | 2013-04-26 | | 公開日 | 2013-07-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.195 Å) | | 主引用文献 | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4K85
 
 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 12:0 Ceramide-1-Phosphate (12:0-C1P) | | 分子名称: | (2S,3R,4E)-2-(dodecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | 著者 | Simanshu, D.K, Brown, R.E, Patel, D.J. | | 登録日 | 2013-04-17 | | 公開日 | 2013-07-17 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4K8N
 
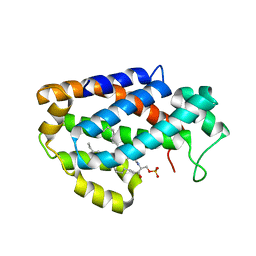 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 18:1 Ceramide-1-Phosphate (18:1-C1P) | | 分子名称: | (2S,3R,4Z)-3-hydroxy-2-[(9E)-octadec-9-enoylamino]octadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | 著者 | Simanshu, D.K, Brown, R.E, Patel, D.J. | | 登録日 | 2013-04-18 | | 公開日 | 2013-07-17 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3.102 Å) | | 主引用文献 | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4IUU
 
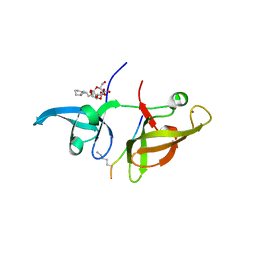 | |
4IUV
 
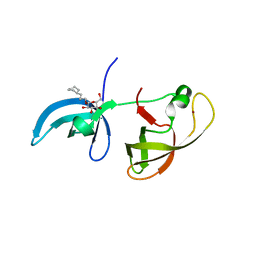 | |
4IUT
 
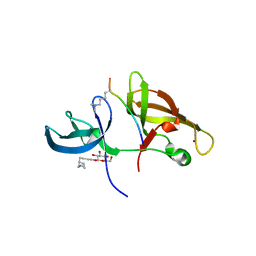 | |
2ASJ
 
 | | oxoG-modified Preinsertion Binary Complex | | 分子名称: | 5'-D(*CP*TP*AP*AP*CP*(8OG)*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*GP*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*(DDG))-3', CALCIUM ION, ... | | 著者 | Rechkoblit, O, Malinina, L, Cheng, Y, Kuryavyi, V, Broyde, S, Geacintov, N.E, Patel, D.J. | | 登録日 | 2005-08-23 | | 公開日 | 2006-01-10 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Stepwise Translocation of Dpo4 Polymerase during Error-Free Bypass of an oxoG Lesion
Plos Biol., 4, 2006
|
|
4JVY
 
 | | Structure of the STAR (signal transduction and activation of RNA) domain of GLD-1 bound to RNA | | 分子名称: | Female germline-specific tumor suppressor gld-1, RNA (5'-R(P*CP*UP*AP*AP*CP*AP*A)-3') | | 著者 | Teplova, M, Hafner, M, Teplov, D, Essig, K, Tuschl, T, Patel, D.J. | | 登録日 | 2013-03-26 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.853 Å) | | 主引用文献 | Structure-function studies of STAR family Quaking proteins bound to their in vivo RNA target sites.
Genes Dev., 27, 2013
|
|
