5U34
 
 | | Crystal structure of AacC2c1-sgRNA binary complex | | Descriptor: | CRISPR-associated endonuclease C2c1, sgRNA | | Authors: | Yang, H, Gao, P, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.255 Å) | | Cite: | PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease.
Cell, 167, 2016
|
|
7CRQ
 
 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (2:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
8T8F
 
 | | Smc5/6 8mer | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 4, Non-structural maintenance of chromosome element 5, ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-22 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular basis for Nse5-6 mediated regulation of Smc5/6 functions.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5VW1
 
 | | Crystal structure of SpyCas9-sgRNA-AcrIIA4 ternary complex | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9/Csn1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, H, Patel, D.J. | | Deposit date: | 2017-05-21 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Inhibition Mechanism of an Anti-CRISPR Suppressor AcrIIA4 Targeting SpyCas9.
Mol. Cell, 67, 2017
|
|
8T8E
 
 | | cryoEM structure of Smc5/6 5mer | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 5, Structural maintenance of chromosomes protein 6 | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-22 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for Nse5-6 mediated regulation of Smc5/6 functions.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
9BI4
 
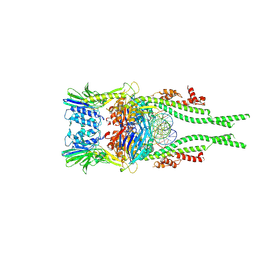 | | cryo EM structure of dsDNA bound Mre11-Rad50 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA repair protein RAD50, Double-strand break repair protein MRE11, ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2024-04-22 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure guided functional analysis of the S. cerevisiae Mre11 complex.
Res Sq, 2024
|
|
9MUD
 
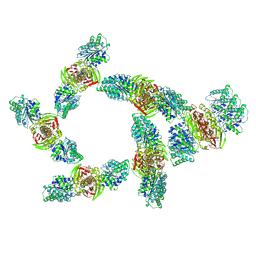 | |
9MUE
 
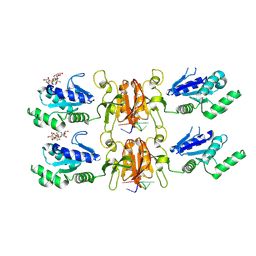 | | Cryo-EM structure of CRISPR-associated cA4 bound Cat1 Pentagonal filament assembly in the presence of NAD (ADPR modelled) | | Descriptor: | Cat1 (CRISPR associated TIR 1) pentagonal filament assembly, RNA (5'-R(P*AP*AP*AP*A)-3'), [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL[HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Majumder, P, Patel, D.J. | | Deposit date: | 2025-01-13 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cat1 forms filament networks to degrade NAD + during the type III CRISPR-Cas antiviral response.
Science, 2025
|
|
9MW9
 
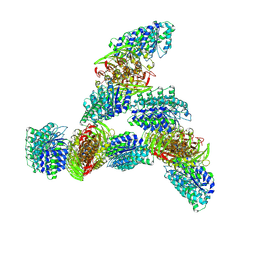 | |
9MUO
 
 | | Cryo-EM structure of CRISPR-associated cA4 bound Cat1 Pentagonal filament assembly in the presence of NAD analog BAD | | Descriptor: | Cat1 (CRISPR-associated TIR 1), RNA (5'-R(P*AP*AP*AP*A)-3'), [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3S,4R,5S)-5-(3-carbamoylphenyl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name) | | Authors: | Majumder, P, Patel, D.J. | | Deposit date: | 2025-01-14 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cat1 forms filament networks to degrade NAD + during the type III CRISPR-Cas antiviral response.
Science, 2025
|
|
4LOI
 
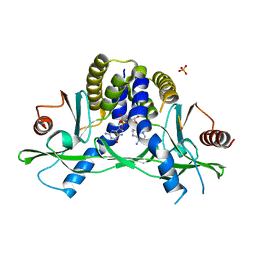 | | Crystal structure of hSTING(H232) in complex with c[G(2',5')pA(2',5')p] | | Descriptor: | 2-amino-9-[(1R,3R,6R,8R,9R,11S,14R,16R,17R,18R)-16-(6-amino-9H-purin-9-yl)-3,11,17,18-tetrahydroxy-3,11-dioxido-2,4,7,10,12,15-hexaoxa-3,11-diphosphatricyclo[12.2.1.1~6,9~]octadec-8-yl]-1,9-dihydro-6H-purin-6-one, PHOSPHATE ION, Stimulator of interferon genes protein | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
4LOL
 
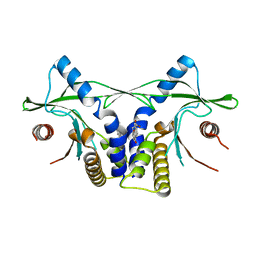 | | Crystal structure of mSting in complex with DMXAA | | Descriptor: | (5,6-dimethyl-9-oxo-9H-xanthen-4-yl)acetic acid, Stimulator of interferon genes protein | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
6UFJ
 
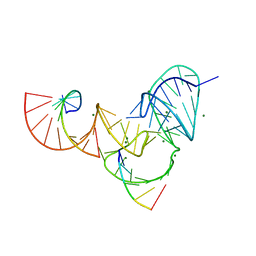 | | Pistol ribozyme product crystal structure | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*UP*CP*CP*AP*G)-3'), RNA (5'-R(*UP*CP*UP*GP*CP*UP*CP*UP*CP*(23G))-3'), ... | | Authors: | Teplova, M, Falschlunger, C, Krasheninina, O, Patel, D.J, Micura, R. | | Deposit date: | 2019-09-24 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Crucial Roles of Two Hydrated Mg2+Ions in Reaction Catalysis of the Pistol Ribozyme.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5K7E
 
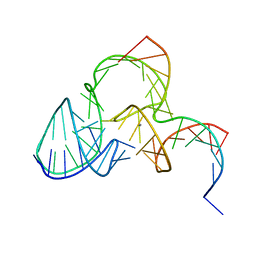 | | The structure of pistol ribozyme, soaked with Mn2+ | | Descriptor: | DNA/RNA 11-MER, MANGANESE (II) ION, RNA 47-MER | | Authors: | Ren, A, Patel, D. | | Deposit date: | 2016-05-26 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Pistol ribozyme adopts a pseudoknot fold facilitating site-specific in-line cleavage.
Nat.Chem.Biol., 12, 2016
|
|
8T66
 
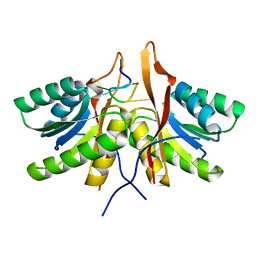 | | cA6 bound Cam1 | | Descriptor: | Cam1, RNA (5'-R(P*AP*AP*AP*AP*A)-3') | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
8T65
 
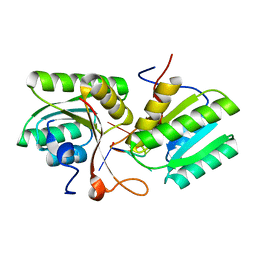 | | cA4 bound Cam1 | | Descriptor: | Cam1, RNA (5'-R(P*AP*AP*AP*A)-3') | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
8T64
 
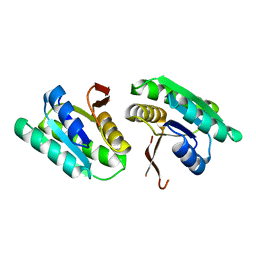 | | Apo Cam1(42-206) | | Descriptor: | Cam1 | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
6O6S
 
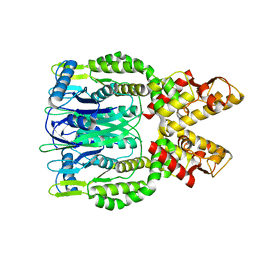 | | Crystal structure of Apo Csm6 | | Descriptor: | Csm6 | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | CRISPR-Cas III-A Csm6 CARF Domain Is a Ring Nuclease Triggering Stepwise cA4Cleavage with ApA>p Formation Terminating RNase Activity.
Mol.Cell, 75, 2019
|
|
5U30
 
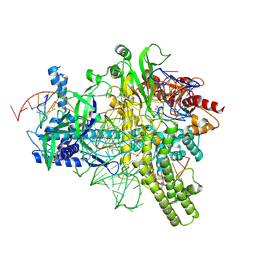 | | Crystal structure of AacC2c1-sgRNA-extended target DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease C2c1, Non-target DNA strand, SULFATE ION, ... | | Authors: | Yang, H, Gao, P, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease.
Cell, 167, 2016
|
|
7SDE
 
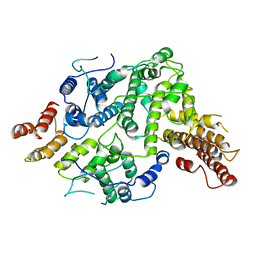 | | Cryo-EM structure of Nse5/6 heterodimer | | Descriptor: | Non-structural maintenance of chromosome element 5, Ubiquitin-like protein SMT3,DNA repair protein KRE29 chimera | | Authors: | Yu, Y, Patel, D.J, Zhao, X.L. | | Deposit date: | 2021-09-29 | | Release date: | 2021-10-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The cryo-EM structure of Nse5/6 complex with the C terminal part of Nse5
To Be Published
|
|
5K7C
 
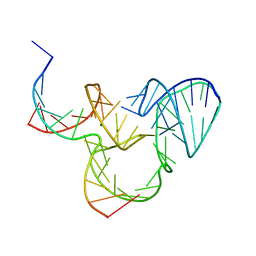 | | The native structure of native pistol ribozyme | | Descriptor: | DNA/RNA 11-MER, MAGNESIUM ION, RNA 47-MER | | Authors: | Ren, A, Patel, D. | | Deposit date: | 2016-05-26 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Pistol ribozyme adopts a pseudoknot fold facilitating site-specific in-line cleavage.
Nat.Chem.Biol., 12, 2016
|
|
5U33
 
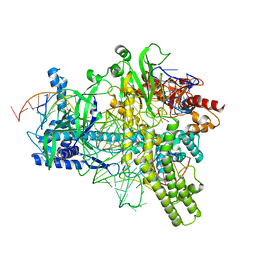 | | Crystal structure of AacC2c1-sgRNA-extended non-target DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease C2c1, Non-target DNA strand, SULFATE ION, ... | | Authors: | Yang, H, Gao, P, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease.
Cell, 167, 2016
|
|
5K7D
 
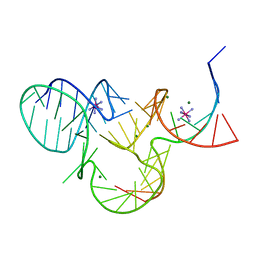 | | The structure of native pistol ribozyme, bound to Iridium | | Descriptor: | DNA/RNA 11-MER, IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Ren, A, Patel, D. | | Deposit date: | 2016-05-26 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Pistol ribozyme adopts a pseudoknot fold facilitating site-specific in-line cleavage.
Nat.Chem.Biol., 12, 2016
|
|
6PPR
 
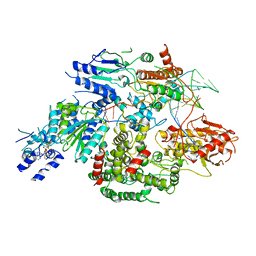 | | Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP and DNA | | Descriptor: | ATP-dependent DNA helicase (UvrD/REP), DNA (70-MER), IRON/SULFUR CLUSTER, ... | | Authors: | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | Deposit date: | 2019-07-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6PPJ
 
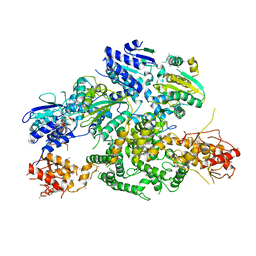 | | Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP | | Descriptor: | ATP-dependent DNA helicase (UvrD/REP), IRON/SULFUR CLUSTER, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | Deposit date: | 2019-07-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
