7S68
 
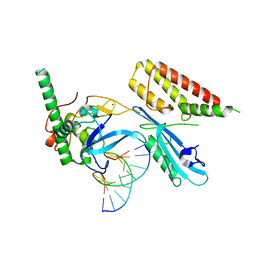 | | Structure of human PARP1 domains (Zn1, Zn3, WGR and HD) bound to a DNA double strand break. | | Descriptor: | DNA (5'-D(*GP*CP*CP*TP*GP*CP*AP*GP*GP*C)-3'), Fusion of PARP1 zinc fingers 1 and 3 (Zn1, Zn3), ... | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
4QNH
 
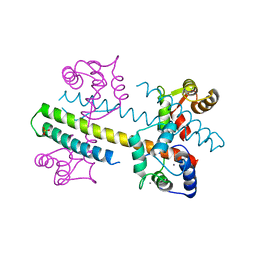 | | Calcium-calmodulin (T79D) complexed with the calmodulin binding domain from a small conductance potassium channel SK2-a | | Descriptor: | CALCIUM ION, Calmodulin, SULFATE ION, ... | | Authors: | Zhang, M, Pascal, J.M, Logothetis, D.E, Zhang, J.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Selective phosphorylation modulates the PIP2 sensitivity of the CaM-SK channel complex.
Nat.Chem.Biol., 10, 2014
|
|
5DS3
 
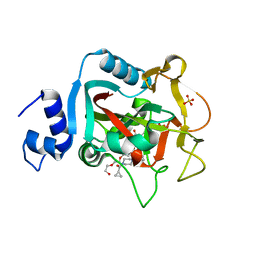 | | Crystal structure of constitutively active PARP-1 | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, PENTAETHYLENE GLYCOL, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Langelier, M.F, Pascal, J.M. | | Deposit date: | 2015-09-16 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | PARP-1 Activation Requires Local Unfolding of an Autoinhibitory Domain.
Mol.Cell, 60, 2015
|
|
5DSY
 
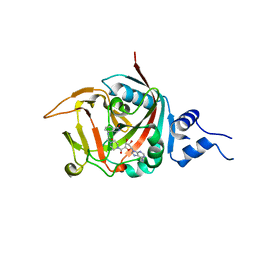 | | Crystal structure of constitutively active PARP-2 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Poly [ADP-ribose] polymerase 2 | | Authors: | Riccio, A.A, Pascal, J.M. | | Deposit date: | 2015-09-17 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | PARP-1 Activation Requires Local Unfolding of an Autoinhibitory Domain.
Mol.Cell, 60, 2015
|
|
7SZ2
 
 | | Mouse PARP13/ZAP ZnF5-WWE1-WWE2 bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ayanath Kuttiyatveetil, J.R, Pascal, J.M. | | Deposit date: | 2021-11-25 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and functional analysis of the ZnF5-WWE1-WWE2 region of PARP13/ZAP define a distinctive mode of engaging poly(ADP-ribose).
Cell Rep, 41, 2022
|
|
7SZ3
 
 | | Mouse PARP13/ZAP ZnF5-WWE1-WWE2 bound to ADPr | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ayanath Kuttiyatveetil, J.R, Pascal, J.M. | | Deposit date: | 2021-11-25 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and functional analysis of the ZnF5-WWE1-WWE2 region of PARP13/ZAP define a distinctive mode of engaging poly(ADP-ribose).
Cell Rep, 41, 2022
|
|
8FYZ
 
 | | Crystal structure of human PARP1 ART domain bound to inhibitor UKTT10 (compound 13) | | Descriptor: | (2P)-2-{3-[(4R)-3-(trifluoromethyl)-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazine-7(8H)-carbonyl]phenyl}-1H-benzimidazole-4-carboxamide, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2023-01-27 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Novel modifications of PARP inhibitor veliparib increase PARP1 binding to DNA breaks.
Biochem.J., 481, 2024
|
|
8FYY
 
 | |
8G0H
 
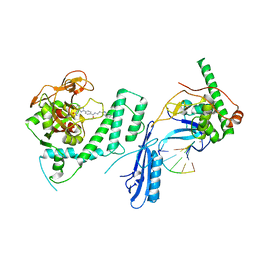 | |
8FZ1
 
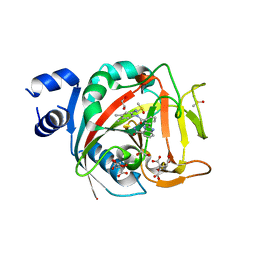 | | Crystal structure of human PARP1 ART domain bound to inhibitor UKTT22 (compound 14) | | Descriptor: | (2P)-2-{3-[(2-amino-4,5-dimethylphenyl)carbamoyl]phenyl}-1H-benzimidazole-4-carboxamide, 1,2-ETHANEDIOL, CITRIC ACID, ... | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2023-01-27 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Novel modifications of PARP inhibitor veliparib increase PARP1 binding to DNA breaks.
Biochem.J., 481, 2024
|
|
4DQY
 
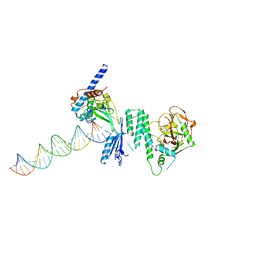 | |
5JHQ
 
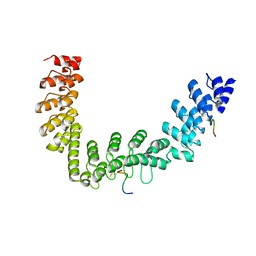 | |
5D5K
 
 | |
3SJQ
 
 | | Crystal structure of a small conductance potassium channel splice variant complexed with calcium-calmodulin | | Descriptor: | 1-phenylurea, CALCIUM ION, Calmodulin, ... | | Authors: | Zhang, M, Pascal, J.M, Zhang, J.-F. | | Deposit date: | 2011-06-21 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for calmodulin as a dynamic calcium sensor.
Structure, 20, 2012
|
|
4J9Y
 
 | | Calcium-calmodulin complexed with the calmodulin binding domain from a small conductance potassium channel splice variant | | Descriptor: | CALCIUM ION, Calmodulin, GLYCEROL, ... | | Authors: | Zhang, M, Pascal, J.M, Zhang, J.-F. | | Deposit date: | 2013-02-17 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Unstructured to structured transition of an intrinsically disordered protein peptide in coupling Ca2+-sensing and SK channel activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4J9Z
 
 | | Calcium-calmodulin complexed with the calmodulin binding domain from a small conductance potassium channel splice variant and NS309 | | Descriptor: | (3E)-6,7-dichloro-3-(hydroxyimino)-1,3-dihydro-2H-indol-2-one, CALCIUM ION, Calmodulin, ... | | Authors: | Zhang, M, Pascal, J.M, Zhang, J.-F. | | Deposit date: | 2013-02-17 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Unstructured to structured transition of an intrinsically disordered protein peptide in coupling Ca2+-sensing and SK channel activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8SWZ
 
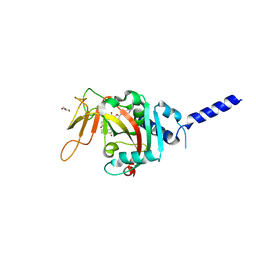 | | PARP4 ART domain bound to EB47 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, GLYCEROL, Protein mono-ADP-ribosyltransferase PARP4 | | Authors: | Frigon, L, Pascal, J.M. | | Deposit date: | 2023-05-19 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical analysis of the PARP1-homology region of PARP4/vault PARP.
Nucleic Acids Res., 51, 2023
|
|
8SX2
 
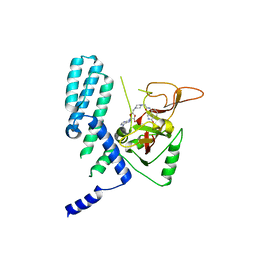 | | PARP4 catalytic domain bound to EB47 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Protein mono-ADP-ribosyltransferase PARP4 | | Authors: | Frigon, L, Pascal, J.M. | | Deposit date: | 2023-05-19 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural and biochemical analysis of the PARP1-homology region of PARP4/vault PARP.
Nucleic Acids Res., 51, 2023
|
|
8SX1
 
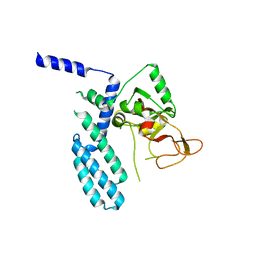 | | PARP4 catalytic domain | | Descriptor: | Protein mono-ADP-ribosyltransferase PARP4 | | Authors: | Frigon, L, Pascal, J.M. | | Deposit date: | 2023-05-19 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural and biochemical analysis of the PARP1-homology region of PARP4/vault PARP.
Nucleic Acids Res., 51, 2023
|
|
8SWY
 
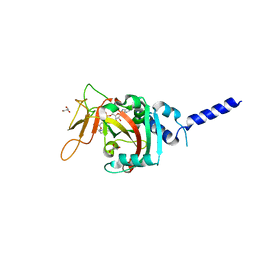 | | PARP4 ART domain bound to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, Protein mono-ADP-ribosyltransferase PARP4 | | Authors: | Frigon, L, Pascal, J.M. | | Deposit date: | 2023-05-19 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and biochemical analysis of the PARP1-homology region of PARP4/vault PARP.
Nucleic Acids Res., 51, 2023
|
|
1OBT
 
 | | STRUCTURE OF RICIN A CHAIN MUTANT, COMPLEX WITH AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, RICIN A CHAIN | | Authors: | Day, P.J, Ernst, S.R, Frankel, A.E, Monzingo, A.F, Pascal, J.M, Svinth, M, Robertus, J.D. | | Deposit date: | 1996-06-22 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and activity of an active site substitution of ricin A chain.
Biochemistry, 35, 1996
|
|
1OBS
 
 | | STRUCTURE OF RICIN A CHAIN MUTANT | | Descriptor: | RICIN A CHAIN | | Authors: | Day, P.J, Ernst, S.R, Frankel, A.E, Monzingo, A.F, Pascal, J.M, Svinth, M, Robertus, J.D. | | Deposit date: | 1996-06-25 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and activity of an active site substitution of ricin A chain.
Biochemistry, 35, 1996
|
|
6NRH
 
 | | Crystal Structure of human PARP-1 ART domain bound inhibitor UTT63 | | Descriptor: | 3-hydroxy-2-({4-[4-(pyrimidin-2-yl)piperazine-1-carbonyl]phenyl}methyl)-1-benzofuran-7-carboxamide, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Langelier, M.F, Pascal, J.M. | | Deposit date: | 2019-01-23 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design and Synthesis of Poly(ADP-ribose) Polymerase Inhibitors: Impact of Adenosine Pocket-Binding Motif Appendage to the 3-Oxo-2,3-dihydrobenzofuran-7-carboxamide on Potency and Selectivity.
J.Med.Chem., 62, 2019
|
|
6NRJ
 
 | | Crystal Structure of human PARP-1 ART domain bound to inhibitor UTT93 | | Descriptor: | (2Z)-2-[(4-{[2-(1H-benzimidazol-2-yl)ethyl]carbamoyl}phenyl)methylidene]-3-oxo-2,3-dihydro-1-benzofuran-7-carboxamide, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Langelier, M.F, Pascal, J.M. | | Deposit date: | 2019-01-23 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design and Synthesis of Poly(ADP-ribose) Polymerase Inhibitors: Impact of Adenosine Pocket-Binding Motif Appendage to the 3-Oxo-2,3-dihydrobenzofuran-7-carboxamide on Potency and Selectivity.
J.Med.Chem., 62, 2019
|
|
6NTU
 
 | |
