6ABA
 
 | | The crystal structure of the photoactivated state of Nonlabens marinus Rhodopsin 3 | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.-H, Ohki, M, Park, J.-H, Jin, Z, Lee, W, Liu, H, Tame, J.R.H, Shibayama, N, Park, S.-Y. | | Deposit date: | 2018-07-20 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | The pumping mechanism of NM-R3, a light-driven marine bacterial chloride importer in the rhodopsin family
To Be Published
|
|
4DFW
 
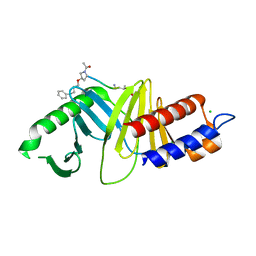 | | Oxime-based Post Solid-phase Peptide Diversification: Identification of High Affinity Polo-like Kinase 1 (Plk1) Polo-box Domain Binding Peptides | | Descriptor: | CHLORIDE ION, Peptide, Serine/threonine-protein kinase PLK1 | | Authors: | Liu, F, Park, J.-E, Qian, W.-J, Lim, D, Gr ber, M, Berg, T, Yaffe, M.B, Lee, K.S, Burke Jr, T.R. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Oxime-based Post Solid-phase Peptide Diversification: Identification of High Affinity Polo-like Kinase 1 (Plk1) Polo-box Domain Binding Peptides
Chem.Biol., 2012
|
|
4HVZ
 
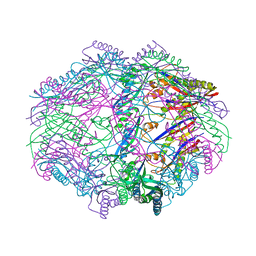 | |
1YAK
 
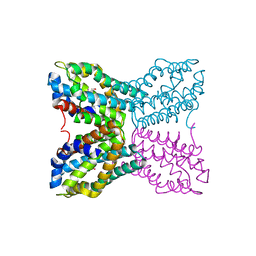 | | Complex of Bacillus subtilis TenA with 4-amino-2-methyl-5-hydroxymethylpyrimidine | | Descriptor: | 4-AMINO-5-HYDROXYMETHYL-2-METHYLPYRIMIDINE, Transcriptional activator tenA | | Authors: | Toms, A.V, Haas, A.L, Park, J.-H, Begley, T.P, Ealick, S.E. | | Deposit date: | 2004-12-17 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural characterization of the regulatory proteins TenA and TenI from Bacillus subtilis and identification of TenA as a thiaminase II.
Biochemistry, 44, 2005
|
|
1YAD
 
 | | Structure of TenI from Bacillus subtilis | | Descriptor: | Regulatory protein tenI, SULFATE ION, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM | | Authors: | Toms, A.V, Haas, A.L, Park, J.-H, Begley, T.P, Ealick, S.E. | | Deposit date: | 2004-12-17 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the regulatory proteins TenA and TenI from Bacillus subtilis and identification of TenA as a thiaminase II.
Biochemistry, 44, 2005
|
|
2X7O
 
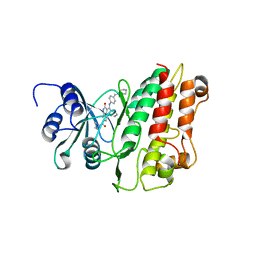 | | Crystal structure of TGFbRI complexed with an indolinone inhibitor | | Descriptor: | (3Z)-N-ETHYL-N-METHYL-2-OXO-3-(PHENYL{[4-(PIPERIDIN-1-YLMETHYL)PHENYL]AMINO}METHYLIDENE)-2,3-DIHYDRO-1H-INDOLE-6-CARBOXAMIDE, TGF-BETA RECEPTOR TYPE I | | Authors: | Roth, G.J, Heckel, A, Brandl, T, Grauert, M, Hoerer, S, Kley, J.T, Schnapp, G, Baum, P, Mennerich, D, Schnapp, A, Park, J.E. | | Deposit date: | 2010-03-03 | | Release date: | 2010-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Design, Synthesis and Evaluation of Indolinones as Inhibitors of the Transforming Growth Factor Beta Receptor I (Tgfbri)
J.Med.Chem., 53, 2010
|
|
4G4V
 
 | |
4G4Y
 
 | |
4G4X
 
 | | Crystal structure of peptidoglycan-associated lipoprotein from Acinetobacter baumannii | | Descriptor: | D-ALANINE, GLYCEROL, Peptidoglycan-associated lipoprotein, ... | | Authors: | Lee, W.C, Song, J.H, Park, J.S, Kim, H.Y. | | Deposit date: | 2012-07-16 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enantiomer-dependent amino acid binding affinity of OmpA-like domains from Acinetobacter baumannii peptidoglycan-associated lipoprotein and OmpA
To be Published
|
|
4G88
 
 | | Crystal structure of OmpA peptidoglycan-binding domain from Acinetobacter baumannii | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, Outer membrane protein Omp38, S,R MESO-TARTARIC ACID | | Authors: | Lee, W.C, Song, J.H, Park, J.S, Kim, H.Y. | | Deposit date: | 2012-07-22 | | Release date: | 2013-07-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enantiomer-dependent amino acid binding affinity of OmpA-like domains from Acinetobacter baumannii peptidoglycan-associated lipoprotein and OmpA
to be published
|
|
1WS1
 
 | |
4G4Z
 
 | |
7YGW
 
 | | Crystal structure of the Zn2+-bound EFhd1/Swiprosin-2 | | Descriptor: | EF-hand domain-containing protein D1, GLYCEROL, ZINC ION | | Authors: | Mun, S.A, Park, J, Kang, J.Y, Park, T, Jin, M, Yang, J, Eom, S.H. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural and biochemical insights into Zn 2+ -bound EF-hand proteins, EFhd1 and EFhd2.
Iucrj, 10, 2023
|
|
7YGV
 
 | | Crystal structure of the Ca2+-bound EFhd1/Swiprosin-2 | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D1, GLYCEROL, ... | | Authors: | Mun, S.A, Park, J, Kang, J.Y, Park, T, Jin, M, Ynag, J, Eom, S.H. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical insights into Zn 2+ -bound EF-hand proteins, EFhd1 and EFhd2.
Iucrj, 10, 2023
|
|
7YGY
 
 | | Crystal structure of the Zn2+-bound EFhd2/Swiprosin-1 | | Descriptor: | EF-hand domain-containing protein D2, ZINC ION | | Authors: | Mun, S.A, Park, J, Kang, J.Y, Park, T, Jin, M, Yang, J, Eom, S.H. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical insights into Zn 2+ -bound EF-hand proteins, EFhd1 and EFhd2.
Iucrj, 10, 2023
|
|
4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WLJ
 
 | | High resolution crystal structure of human kynurenine aminotransferase-I in complex with aminooxyacetate | | Descriptor: | 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, Kynurenine--oxoglutarate transaminase 1 | | Authors: | Nadvi, N.A, Salam, N.K, Park, J, Akladios, F.N, Kapoor, V, Collyer, C.A, Gorrell, M.D, Church, W.B. | | Deposit date: | 2014-10-07 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | High resolution crystal structures of human kynurenine aminotransferase-I bound to PLP cofactor, and in complex with aminooxyacetate.
Protein Sci., 26, 2017
|
|
4WP0
 
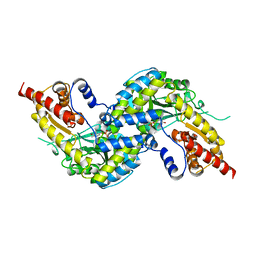 | | Crystal structure of human kynurenine aminotransferase-I with a C-terminal V5-hexahistidine tag | | Descriptor: | Kynurenine--oxoglutarate transaminase 1 | | Authors: | Nadvi, N.A, Salam, N.K, Park, J, Akladios, F.N, Kapoor, V, Collyer, C.A, Gorrell, M.D, Church, W.B. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human kynurenine aminotransferase-I in a novel space group
To be published
|
|
2R64
 
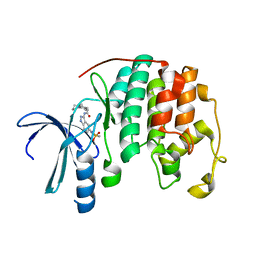 | | Crystal structure of a 3-aminoindazole compound with CDK2 | | Descriptor: | Cell division protein kinase 2, N-[5-(1,1-DIOXIDOISOTHIAZOLIDIN-2-YL)-1H-INDAZOL-3-YL]-2-(4-PIPERIDIN-1-YLPHENYL)ACETAMIDE | | Authors: | Lee, J, Choi, H, Kim, K.H, Jeong, S, Park, J.W, Baek, C.S, Lee, S.H. | | Deposit date: | 2007-09-05 | | Release date: | 2008-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and biological evaluation of 3,5-diaminoindazoles as cyclin-dependent kinase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4WLH
 
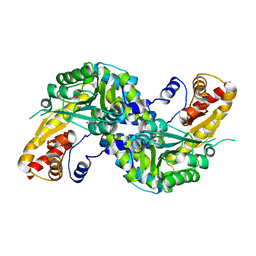 | | High resolution crystal structure of human kynurenine aminotransferase-I bound to PLP cofactor | | Descriptor: | Kynurenine--oxoglutarate transaminase 1 | | Authors: | Nadvi, N.A, Salam, N.K, Park, J, Akladios, F.N, Kapoor, V, Collyer, C.A, Gorrell, M.D, Church, W.B. | | Deposit date: | 2014-10-07 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | High resolution crystal structures of human kynurenine aminotransferase-I bound to PLP cofactor, and in complex with aminooxyacetate.
Protein Sci., 26, 2017
|
|
4ITB
 
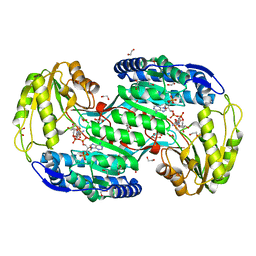 | | Structure of bacterial enzyme in complex with cofactor and substrate | | Descriptor: | 1,2-ETHANEDIOL, 4-oxobutanoic acid, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Rhee, S, Park, J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-24 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis for a Cofactor-dependent Oxidation Protection and Catalysis of Cyanobacterial Succinic Semialdehyde Dehydrogenase.
J.Biol.Chem., 288, 2013
|
|
1YAF
 
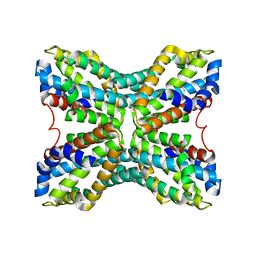 | | Structure of TenA from Bacillus subtilis | | Descriptor: | Transcriptional activator tenA | | Authors: | Toms, A.V, Haas, A.L, Park, J.-H, Begley, T.P, Ealick, S.E. | | Deposit date: | 2004-12-17 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural characterization of the regulatory proteins TenA and TenI from Bacillus subtilis and identification of TenA as a thiaminase II.
Biochemistry, 44, 2005
|
|
4IT9
 
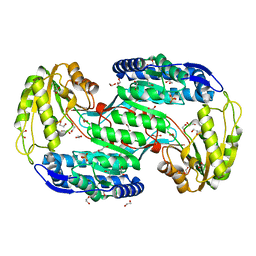 | | Structure of Bacterial Enzyme | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Succinate-semialdehyde dehydrogenase | | Authors: | Rhee, S, Park, J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for a Cofactor-dependent Oxidation Protection and Catalysis of Cyanobacterial Succinic Semialdehyde Dehydrogenase.
J.Biol.Chem., 288, 2013
|
|
4ITA
 
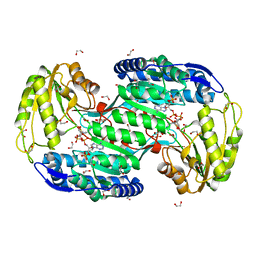 | | Structure of bacterial enzyme in complex with cofactor | | Descriptor: | 1,2-ETHANEDIOL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Succinate-semialdehyde dehydrogenase | | Authors: | Rhee, S, Park, J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-24 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis for a Cofactor-dependent Oxidation Protection and Catalysis of Cyanobacterial Succinic Semialdehyde Dehydrogenase.
J.Biol.Chem., 288, 2013
|
|
1WS0
 
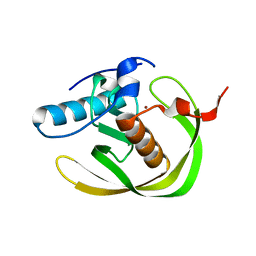 | |
