3ULB
 
 | |
3VOQ
 
 | |
3W15
 
 | | Structure of peroxisomal targeting signal 2 (PTS2) of Saccharomyces cerevisiae 3-ketoacyl-CoA thiolase in complex with Pex7p and Pex21p | | Descriptor: | 3-ketoacyl-CoA thiolase, peroxisomal, Maltose-binding periplasmic protein, ... | | Authors: | Pan, D, Nakatsu, T, Kato, H. | | Deposit date: | 2012-11-06 | | Release date: | 2013-07-03 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of peroxisomal targeting signal-2 bound to its receptor complex Pex7p-Pex21p
Nat.Struct.Mol.Biol., 20, 2013
|
|
3ULC
 
 | |
7FC9
 
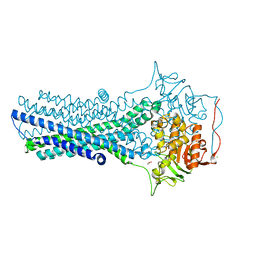 | | Crystal structure of CmABCB1 in lipidic mesophase revealed by LCP-SFX | | Descriptor: | ACETATE ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Pan, D, Oyama, R, Sato, T, Nakane, T, Mizunuma, R, Matsuoka, K, Joti, Y, Tono, K, Nango, E, Iwata, S, Nakatsu, T, Kato, H. | | Deposit date: | 2021-07-14 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of CmABCB1 multi-drug exporter in lipidic mesophase revealed by LCP-SFX.
Iucrj, 9, 2022
|
|
6EUM
 
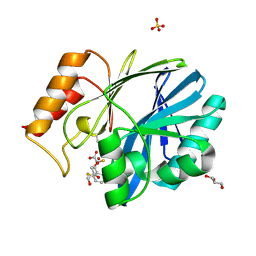 | | CRYSTAL STRUCTURE OF BCII METALLO-BETA-LACTAMASE IN COMPLEX WITH DZ-307 | | Descriptor: | (~{Z})-2-sulfanyl-3-[2,3,6-tris(fluoranyl)phenyl]prop-2-enoic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Stepanovs, D, McDonough, M.A, Schofield, C.J, Zhang, D, El-Husseiny, A, Brem, J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structure activity relationship studies on rhodanines and derived enethiol inhibitors of metallo-beta-lactamases.
Bioorg. Med. Chem., 26, 2018
|
|
6EWE
 
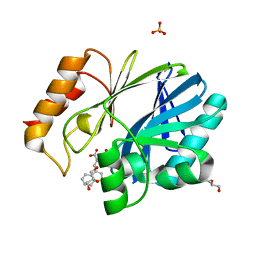 | | Crystal structure of BCII Metallo-beta-lactamase in complex with DZ-308 | | Descriptor: | (~{Z})-3-(1-benzothiophen-3-yl)-2-sulfanyl-prop-2-enoic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Stepanovs, D, McDonough, M.A, Schofield, C.J, Zhang, D, El-Husseiny, A, Brem, J. | | Deposit date: | 2017-11-03 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure activity relationship studies on rhodanines and derived enethiol inhibitors of metallo-beta-lactamases.
Bioorg. Med. Chem., 26, 2018
|
|
4IWQ
 
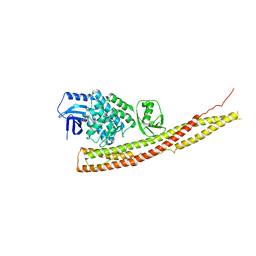 | | Crystal structure and mechanism of activation of TBK1 | | Descriptor: | N-{3-[(5-cyclopropyl-2-{[3-(morpholin-4-ylmethyl)phenyl]amino}pyrimidin-4-yl)amino]propyl}cyclobutanecarboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Larabi, A. | | Deposit date: | 2013-01-24 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure and mechanism of activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
4IWO
 
 | | Crystal structure and mechanism of activation of TBK1 | | Descriptor: | N-{3-[(5-cyclopropyl-2-{[3-(2-oxopyrrolidin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]propyl}cyclobutanecarboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Larabi, A. | | Deposit date: | 2013-01-24 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure and mechanism of activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
4IWP
 
 | | Crystal structure and mechanism of activation of TBK1 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Larabi, A. | | Deposit date: | 2013-01-24 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.065 Å) | | Cite: | Crystal structure and mechanism of activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
2O6G
 
 | |
6RSU
 
 | | TBK1 in complex with Inhibitor compound 35 | | Descriptor: | 3,3,3-tris(fluoranyl)-1-[4-[(1~{R})-1-[2-[[(2~{S})-5-(5-propan-2-yloxypyrimidin-4-yl)-2,3-dihydro-1~{H}-benzimidazol-2-yl]amino]pyridin-4-yl]ethyl]piperazin-1-yl]propan-1-one, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Hillig, R.C, Rengachari, S. | | Deposit date: | 2019-05-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of BAY-985, a Highly Selective TBK1/IKK epsilon Inhibitor.
J.Med.Chem., 63, 2020
|
|
6RSR
 
 | | TBK1 in complex with compound 2 | | Descriptor: | Serine/threonine-protein kinase TBK1, ~{N}-(cyclopropen-1-ylmethyl)-2-[[4-[[4-[3,3,3-tris(fluoranyl)propanoyl]piperazin-1-yl]methyl]pyridin-2-yl]amino]-1~{H}-benzimidazole-5-carboxamide | | Authors: | Panne, D, Hillig, R.C, Rengachari, S. | | Deposit date: | 2019-05-22 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Discovery of BAY-985, a Highly Selective TBK1/IKK epsilon Inhibitor.
J.Med.Chem., 63, 2020
|
|
6RST
 
 | | TBK1 in complex with inhibitor compound 24 | | Descriptor: | 1-[4-[(1~{R})-1-[2-[[5-[1-(cyclopropylmethyl)pyrazol-4-yl]-1~{H}-benzimidazol-2-yl]amino]pyridin-4-yl]ethyl]piperazin-1-yl]-3,3,3-tris(fluoranyl)propan-1-one, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Hillig, R.C, Rengachari, S. | | Deposit date: | 2019-05-22 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Discovery of BAY-985, a Highly Selective TBK1/IKK epsilon Inhibitor.
J.Med.Chem., 63, 2020
|
|
6QPW
 
 | | Structural basis of cohesin ring opening | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Sister chromatid cohesion protein 1, ... | | Authors: | Panne, D, Muir, K.W, Li, Y, Weis, F. | | Deposit date: | 2019-02-15 | | Release date: | 2020-02-05 | | Last modified: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of the cohesin ATPase elucidates the mechanism of SMC-kleisin ring opening.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5JMX
 
 | | Crystal Structure of BcII metallo-beta-lactamase in complex with DZ-305 | | Descriptor: | (2Z)-3-(4-fluorophenyl)-2-sulfanylprop-2-enoic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Stepanovs, D, McDonough, M.A, Schofield, C.J, Zhang, D, Brem, J. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure activity relationship studies on rhodanines and derived enethiol inhibitors of metallo-beta-lactamases.
Bioorg. Med. Chem., 26, 2018
|
|
8JQU
 
 | | Crystal structure of GppNHp bound GTPase domain of Rab5a from Leishmania donovani | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Pandey, D, Zohib, M, Pal, R.K, Biswal, B.K, Arora, A. | | Deposit date: | 2023-06-14 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Crystal structure of GppNHp bound GTPase domain of Rab5a from Leishmania donovani
To Be Published
|
|
1T2K
 
 | | Structure Of The DNA Binding Domains Of IRF3, ATF-2 and Jun Bound To DNA | | Descriptor: | 31-MER, Cyclic-AMP-dependent transcription factor ATF-2, Interferon regulatory factor 3, ... | | Authors: | Panne, D, Maniatis, T, Harrison, S.C. | | Deposit date: | 2004-04-21 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of ATF-2/c-Jun and IRF-3 bound to the interferon-beta enhancer.
Embo J., 23, 2004
|
|
2O61
 
 | |
6GYR
 
 | | Transcription factor dimerization activates the p300 acetyltransferase | | Descriptor: | Histone acetyltransferase p300, ZINC ION, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate | | Authors: | Panne, D, Ortega, E. | | Deposit date: | 2018-07-01 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Transcription factor dimerization activates the p300 acetyltransferase.
Nature, 562, 2018
|
|
6GYT
 
 | |
5D9D
 
 | | Luciferin-regenerating enzyme solved by SAD using synchrotron radiation at room temperature | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION, ... | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography.
Sci Rep, 5, 2015
|
|
5D9C
 
 | | Luciferin-regenerating enzyme solved by SIRAS using XFEL (refined against Hg derivative data) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION, ... | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography.
Sci Rep, 5, 2015
|
|
7AMZ
 
 | | Crystal structure of human Butyrylcholinesterase in complex with N-((2S,3R)-4-((2,2-dimethylpropyl)amino)-3-hydroxy-1-phenylbutan-2-yl)-2,2-diphenylacetamide | | Descriptor: | 2,2-dimethylpropyl-[(2~{R},3~{S})-3-(2,2-diphenylethanoylamino)-2-oxidanyl-4-phenyl-butyl]azanium, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Pasieka, A, Panek, D, Wieckowska, A. | | Deposit date: | 2020-10-10 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of multifunctional anti-Alzheimer's agents with a unique mechanism of action including inhibition of the enzyme butyrylcholinesterase and gamma-aminobutyric acid transporters.
Eur.J.Med.Chem., 218, 2021
|
|
5XFE
 
 | | Luciferin-regenerating enzyme solved by SAD using XFEL (refined against 11,000 patterns) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION, ... | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2017-04-10 | | Release date: | 2017-08-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental phase determination with selenomethionine or mercury-derivatization in serial femtosecond crystallography
IUCrJ, 4, 2017
|
|
