4UN1
 
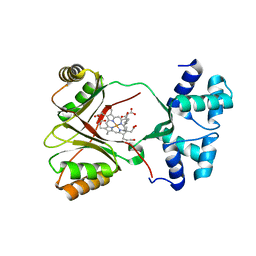 | | Sirohaem decarboxylase AhbA/B - an enzyme with structural homology to the Lrp/AsnC transcription factor family that is part of the alternative haem biosynthesis pathway. | | 分子名称: | 12,18-DIDECARBOXY-SIROHEME, PUTATIVE TRANSCRIPTIONAL REGULATOR, ASNC FAMILY | | 著者 | Palmer, D.J, Brown, D.G, Warren, M.J, Pickersgill, R.W. | | 登録日 | 2014-05-23 | | 公開日 | 2014-06-11 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | The Structure, Function and Properties of Sirohaem Decarboxylase - an Enzyme with Structural Homology to a Transcription Factor Family that is Part of the Alternative Haem Biosynthesis Pathway.
Mol.Microbiol., 93, 2014
|
|
4CZD
 
 | |
3MZ0
 
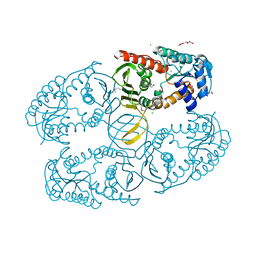 | | Crystal structure of apo myo-inositol dehydrogenase from Bacillus subtilis | | 分子名称: | CHLORIDE ION, GLYCEROL, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, ... | | 著者 | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2010-05-11 | | 公開日 | 2010-09-29 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.539 Å) | | 主引用文献 | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
6TZU
 
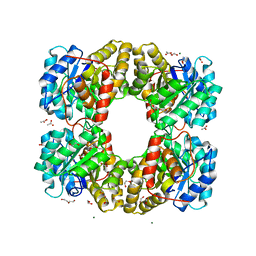 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84A mutant with pyruvate bound in the active site | | 分子名称: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | 著者 | Saran, S, Majdi Yazdi, M, Lehnert, C, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2019-08-13 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J.Struct.Biol., 209, 2020
|
|
6U01
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84D mutant with pyruvate bound in the active site | | 分子名称: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | 著者 | Saran, S, Majdi Yazdi, M, Lehnert, L, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2019-08-13 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J.Struct.Biol., 209, 2020
|
|
5F1V
 
 | | biomimetic design results in a potent allosteric inhibitor of dihydrodipicolinate synthase from Campylobacter jejuni | | 分子名称: | (2R,5R)-2,5-diamino-2,5-bis(4-aminobutyl)hexanedioic acid, 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ... | | 著者 | Conly, C.J.T, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2015-11-30 | | 公開日 | 2016-02-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Biomimetic Design Results in a Potent Allosteric Inhibitor of Dihydrodipicolinate Synthase from Campylobacter jejuni.
J.Am.Chem.Soc., 138, 2016
|
|
5F1U
 
 | | biomimetic design results in a potent allosteric inhibitor of dihydrodipicolinate synthase from Campylobacter jejuni | | 分子名称: | (2R,5R)-2,5-diamino-2,5-bis(4-aminobutyl)hexanedioic acid, 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ... | | 著者 | Conly, C.J.T, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2015-11-30 | | 公開日 | 2016-02-17 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Biomimetic Design Results in a Potent Allosteric Inhibitor of Dihydrodipicolinate Synthase from Campylobacter jejuni.
J.Am.Chem.Soc., 138, 2016
|
|
3VW7
 
 | | Crystal structure of human protease-activated receptor 1 (PAR1) bound with antagonist vorapaxar at 2.2 angstrom | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Proteinase-activated receptor 1, ... | | 著者 | Zhang, C, Srinivasan, Y, Arlow, D.H, Fung, J.J, Palmer, D, Zheng, Y, Green, H.F, Pandey, A, Dror, R.O, Shaw, D.E, Weis, W.I, Coughlin, S.R, Kobilka, B.K. | | 登録日 | 2012-08-07 | | 公開日 | 2012-12-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | High-resolution crystal structure of human protease-activated receptor 1
Nature, 492, 2012
|
|
4K2M
 
 | | Crystal structure of ntda from bacillus subtilis in complex with the plp external aldimine adduct with kanosamine-6-phosphate | | 分子名称: | 1,2-ETHANEDIOL, 3-deoxy-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-O-phosphono-alpha-D-gluco pyranose, ACETATE ION, ... | | 著者 | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2013-04-09 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | The Structure of NtdA, a Sugar Aminotransferase Involved in the Kanosamine Biosynthetic Pathway in Bacillus subtilis, Reveals a New Subclass of Aminotransferases.
J.Biol.Chem., 288, 2013
|
|
4K2B
 
 | |
4K2I
 
 | | Crystal structure of ntda from bacillus subtilis with bound cofactor pmp | | 分子名称: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ACETATE ION, ... | | 著者 | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2013-04-09 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.225 Å) | | 主引用文献 | The Structure of NtdA, a Sugar Aminotransferase Involved in the Kanosamine Biosynthetic Pathway in Bacillus subtilis, Reveals a New Subclass of Aminotransferases.
J.Biol.Chem., 288, 2013
|
|
4L8V
 
 | | Crystal Structure of A12K/D35S mutant myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NADP | | 分子名称: | 1,2-ETHANEDIOL, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Bertwistle, D, Sanders, D.A.R, Palmer, D.R.J. | | 登録日 | 2013-06-18 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Converting NAD-Specific Inositol Dehydrogenase to an Efficient NADP-Selective Catalyst, with a Surprising Twist.
Biochemistry, 52, 2013
|
|
4L9R
 
 | |
1BQG
 
 | | THE STRUCTURE OF THE D-GLUCARATE DEHYDRATASE PROTEIN FROM PSEUDOMONAS PUTIDA | | 分子名称: | D-GLUCARATE DEHYDRATASE | | 著者 | Gulick, A.M, Palmer, D.R.J, Babbitt, P.C, Gerlt, J.A, Rayment, I. | | 登録日 | 1998-08-15 | | 公開日 | 1999-05-18 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Evolution of enzymatic activities in the enolase superfamily: crystal structure of (D)-glucarate dehydratase from Pseudomonas putida.
Biochemistry, 37, 1998
|
|
7KZ6
 
 | | Crystal structure of KabA from Bacillus cereus UW85 with bound cofactor PMP | | 分子名称: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme | | 著者 | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2020-12-10 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
7KZ3
 
 | | Crystal structure of KabA from Bacillus cereus UW85 in complex with the internal aldimine | | 分子名称: | 1,2-ETHANEDIOL, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, SODIUM ION | | 著者 | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2020-12-09 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
7KZ5
 
 | | Crystal structure of KabA from Bacillus cereus UW85 in complex with the plp external aldimine adduct with kanosamine-6-phosphate | | 分子名称: | 1,2-ETHANEDIOL, 3-deoxy-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-O-phosphono-alpha-D-gluco pyranose, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, ... | | 著者 | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2020-12-10 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
7KZD
 
 | | Crystal structure of KabA from Bacillus cereus UW85 in complex with the reduced internal aldimine and with bound Glutarate | | 分子名称: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 1,2-ETHANEDIOL, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, ... | | 著者 | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2020-12-10 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
2VC6
 
 | | Structure of MosA from S. meliloti with pyruvate bound | | 分子名称: | DIHYDRODIPICOLINATE SYNTHASE | | 著者 | Phenix, C.P, Nienaber, K.H, Tam, P.H, Delbaere, L.T.J, Palmer, D.R.J. | | 登録日 | 2007-09-18 | | 公開日 | 2008-06-17 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural, functional and calorimetric investigation of MosA, a dihydrodipicolinate synthase from Sinorhizobium meliloti l5-30, does not support involvement in rhizopine biosynthesis.
Chembiochem, 9, 2008
|
|
3NT2
 
 | | Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2010-07-02 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3003 Å) | | 主引用文献 | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
3NT4
 
 | | Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NADH and inositol | | 分子名称: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase | | 著者 | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2010-07-02 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5001 Å) | | 主引用文献 | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
3NTQ
 
 | |
3NTR
 
 | | Crystal structure of K97V mutant of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NAD and inositol | | 分子名称: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2010-07-05 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.6503 Å) | | 主引用文献 | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
3NT5
 
 | | Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor and product inosose | | 分子名称: | (2R,3S,4s,5R,6S)-2,3,4,5,6-pentahydroxycyclohexanone, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, ... | | 著者 | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2010-07-02 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.9006 Å) | | 主引用文献 | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
3NTO
 
 | |
