6FM8
 
 | | RPAP3 c-terminus | | Descriptor: | RNA polymerase II-associated protein 3 | | Authors: | Pal, M, Roe, S.M. | | Deposit date: | 2018-01-30 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | RPAP3 provides a flexible scaffold for coupling HSP90 to the human R2TP co-chaperone complex
Nat Commun, 2020
|
|
7OLE
 
 | | Cryo-EM structure of the TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2, ... | | Authors: | Pal, M, Llorca, O, Pearl, L. | | Deposit date: | 2021-05-19 | | Release date: | 2021-07-07 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structure of the TELO2-TTI1-TTI2 complex and its function in TOR recruitment to the R2TP chaperone.
Cell Rep, 36, 2021
|
|
3NFT
 
 | | Near-atomic resolution analysis of BipD- A component of the type-III secretion system of Burkholderia pseudomallei | | Descriptor: | Translocator protein bipD | | Authors: | Pal, M, Erskine, P.T, Gill, R.S, Wood, S.P, Cooper, J.B. | | Deposit date: | 2010-06-10 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Near-atomic resolution analysis of BipD, a component of the type III secretion system of Burkholderia pseudomallei.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7SA1
 
 | | LRR-F-Box plant ubiquitin ligase | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, F-box/LRR-repeat MAX2 homolog, ... | | Authors: | Palayam, M, Shabek, N. | | Deposit date: | 2021-09-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | A conformational switch in the SCF-D3/MAX2 ubiquitin ligase facilitates strigolactone signalling.
Nat.Plants, 8, 2022
|
|
6X24
 
 | | Structural basis of plant blue light photoreceptor | | Descriptor: | Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Palayam, M, Ganapathy, J, Guercio, M.A, Shabek, N. | | Deposit date: | 2020-05-19 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural insights into photoactivation of plant Cryptochrome-2.
Commun Biol, 4, 2021
|
|
7UPV
 
 | |
5WVX
 
 | | Crystal Structure of bifunctional Kunitz type Trypsin /amylase inhibitor (AMTIN) from the tubers of Alocasia macrorrhiza | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CITRIC ACID, Trypsin/chymotrypsin inhibitor | | Authors: | Palayam, M, Radhakrishnan, M, Lakshminarayanan, K, Balu, K.E, Ganapathy, J, Krishnasamy, G. | | Deposit date: | 2016-12-29 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural insights into a multifunctional inhibitor, 'AMTIN' from tubers of Alocasia macrorrhizos and its possible role in dengue protease (NS2B-NS3) inhibition.
Int. J. Biol. Macromol., 113, 2018
|
|
5HHD
 
 | | Crystal Structure of Chemically Synthesized Heterochiral {RFX037 plus VEGF-A} Protein Complex in space group P21 | | Descriptor: | D-Peptide RFX037.D, D-Vascular endothelial growth factor, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Uppalapati, M, LEE, D.J, Mandal, K, Kent, S.B.H, Sidhu, S. | | Deposit date: | 2016-01-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent d-Protein Antagonist of VEGF-A is Nonimmunogenic, Metabolically Stable, and Longer-Circulating in Vivo.
Acs Chem.Biol., 11, 2016
|
|
5KKV
 
 | |
5HHC
 
 | | Crystal Structure of Chemically Synthesized Heterochiral {RFX037 plus VEGF-A} Protein Complex in space group P21/n | | Descriptor: | D- Vascular endothelial growth factor-A, GLYCEROL, Vascular endothelial growth factor A | | Authors: | Uppalapati, M, Lee, D.J, Mandal, K, Kent, S.B.H, Sidhu, S. | | Deposit date: | 2016-01-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent d-Protein Antagonist of VEGF-A is Nonimmunogenic, Metabolically Stable, and Longer-Circulating in Vivo.
Acs Chem.Biol., 11, 2016
|
|
8VCE
 
 | | Crystal Structure of plant Carboxylesterase 20 | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Probable carboxylesterase 120 | | Authors: | Palayam, M, Shabek, N. | | Deposit date: | 2023-12-14 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into strigolactone catabolism by carboxylesterases reveal a conserved conformational regulation.
Nat Commun, 15, 2024
|
|
8VCD
 
 | |
8VCA
 
 | | Crystal Structure of plant Carboxylesterase 15 | | Descriptor: | Carboxylesterase 15, GLYCEROL | | Authors: | Palayam, M, Shabek, N. | | Deposit date: | 2023-12-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into strigolactone catabolism by carboxylesterases reveal a conserved conformational regulation.
Nat Commun, 15, 2024
|
|
7EH3
 
 | |
4CHH
 
 | | N-terminal domain of yeast PIH1p | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN INTERACTING WITH HSP90 1 | | Authors: | Roe, S.M, Pal, M. | | Deposit date: | 2013-12-02 | | Release date: | 2014-05-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
4CGW
 
 | |
4CGU
 
 | | Full length Tah1 bound to yeast PIH1 and HSP90 peptide SRMEEVD | | Descriptor: | GLYCEROL, HEAT SHOCK PROTEIN HSP 90-ALPHA, PROTEIN INTERACTING WITH HSP90 1, ... | | Authors: | Roe, S.M, Pal, M. | | Deposit date: | 2013-11-26 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
4CGV
 
 | | First TPR of Spaghetti (RPAP3) bound to HSP90 peptide SRMEEVD | | Descriptor: | GLYCEROL, HEAT SHOCK PROTEIN HSP 90-ALPHA, RNA POLYMERASE II-ASSOCIATED PROTEIN 3 | | Authors: | Roe, S.M, Pal, M. | | Deposit date: | 2013-11-26 | | Release date: | 2014-05-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
4CGQ
 
 | | Full length Tah1 bound to HSP90 peptide SRMEEVD | | Descriptor: | CHLORIDE ION, HEAT SHOCK PROTEIN HSP 90-ALPHA, TPR REPEAT-CONTAINING PROTEIN ASSOCIATED WITH HSP90 | | Authors: | Roe, S.M, Pal, M. | | Deposit date: | 2013-11-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tah1 Helix-Swap Dimerization Prevents Mixed Hsp90 Co-Chaperone Complexes
Acta Crystallogr.,Sect.F, 71, 2015
|
|
9G92
 
 | | Crystal structure of thioredoxin reductase from Cryptosporidium parvum in the "in" conformation | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Gabriele, F, Palerma, M, Ardini, M, Bogard, J, Chen, X.M, Williams, D.L, Angelucci, F. | | Deposit date: | 2024-07-24 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Targeting Apicomplexan Parasites: Structural and Functional Characterization of Cryptosporidium Thioredoxin Reductase as a Novel Drug Target.
Biochemistry, 64, 2025
|
|
4GLN
 
 | | Crystal Structure of Chemically Synthesized Heterochiral {D-Protein Antagonist plus VEGF-A} Protein Complex in space group P21/n | | Descriptor: | D-RFX001, Vascular endothelial growth factor A | | Authors: | Mandal, K, Uppalapati, M, Ault-Riche, D, Kenney, J, Lowitz, J, Sidhu, S, Kent, S.B.H. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GLU
 
 | | Crystal structure of the mirror image form of VEGF-A | | Descriptor: | ACETATE ION, D- Vascular endothelial growth factor-A, GLYCEROL, ... | | Authors: | Mandal, K, Uppalapati, M, Ault-Riche, D, Kenney, J, Lowitz, J, Sidhu, S, Kent, S.B.H. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GLS
 
 | | Crystal Structure of Chemically Synthesized Heterochiral {D-Protein Antagonist plus VEGF-A} Protein Complex in space group P21 | | Descriptor: | D- RFX001, D- Vascular endothelial growth factor-A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mandal, K, Uppalapati, M, Ault-Riche, D, Kenney, J, Lowitz, J, Sidhu, S, Kent, S.B.H. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5BNG
 
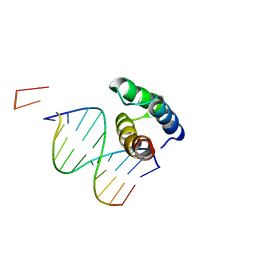 | | monomer of TALE type homeobox transcription factor MEIS1 complexes with specific DNA | | Descriptor: | DNA (5'-D(P*AP*AP*TP*TP*AP*GP*CP*TP*GP*TP*CP*A)-3'), DNA (5'-D(P*TP*GP*AP*CP*AP*GP*CP*TP*AP*A)-3'), DNA (5'-D(P*TP*GP*AP*CP*AP*GP*CP*TP*AP*A-3'), ... | | Authors: | Morgunova, E, Jolma, A, Yin, Y, Nitta, K, Dave, K, Popov, A, Taipale, M, Enge, M, Kivioja, T, Taipale, J. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | DNA-dependent formation of transcription factor pairs alters their binding specificity.
Nature, 527, 2015
|
|
1QSU
 
 | | CRYSTAL STRUCTURE OF THE TRIPLE-HELICAL COLLAGEN-LIKE PEPTIDE, (PRO-HYP-GLY)4-GLU-LYS-GLY(PRO-HYP-GLY)5 | | Descriptor: | PROTEIN ((PRO-HYP-GLY)4- GLU-LYS-GLY(PRO-HYP-GLY)5) | | Authors: | Kramer, R.Z, Venugopal, M, Bella, J, Brodsky, B, Berman, H.M. | | Deposit date: | 1999-06-23 | | Release date: | 2000-08-30 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Staggered molecular packing in crystals of a collagen-like peptide with a single charged pair.
J.Mol.Biol., 301, 2000
|
|
