3KX0
 
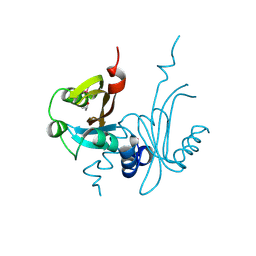 | | Crystal Structure of the PAS domain of Rv1364c | | Descriptor: | ISOPROPYL ALCOHOL, Uncharacterized protein Rv1364c/MT1410 | | Authors: | Jaiswal, R.K, Gopal, B. | | Deposit date: | 2009-12-02 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of a PAS sensor domain in the Mycobacterium tuberculosis transcription regulator Rv1364c
Biochem.Biophys.Res.Commun., 398, 2010
|
|
3KI9
 
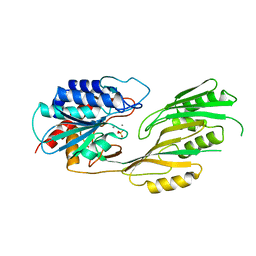 | |
6NET
 
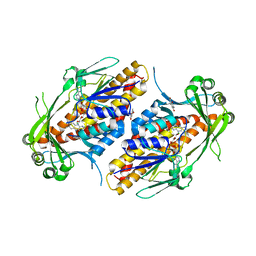 | | FAD-dependent monooxygenase TropB from T. stipitatus substrate complex | | Descriptor: | 2,4-dihydroxy-3,6-dimethylbenzaldehyde, CHLORIDE ION, FAD-dependent monooxygenase tropB, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
6NES
 
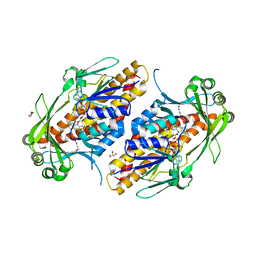 | | FAD-dependent monooxygenase TropB from T. stipitatus | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
3U4C
 
 | |
3VEP
 
 | | Crystal structure of SigD4 in complex with its negative regulator RsdA | | Descriptor: | Probable RNA polymerase sigma-D factor, SULFATE ION, Uncharacterized protein Rv3413c/MT3522 | | Authors: | Jaiswal, R.K, Gopal, B. | | Deposit date: | 2012-01-09 | | Release date: | 2013-02-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis RsdA provides a conformational rationale for selective regulation of sigma-factor activity by proteolysis
Nucleic Acids Res., 41, 2013
|
|
3U4D
 
 | |
3KHZ
 
 | |
3KHX
 
 | |
3U49
 
 | |
4NQW
 
 | |
3S3K
 
 | |
4BXI
 
 | | Crystal structure of ATP binding domain of AgrC from Staphylococcus aureus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACCESSORY GENE REGULATOR PROTEIN C, ACETATE ION, ... | | Authors: | Srivastava, S.K, Rajasree, K, Gopal, B. | | Deposit date: | 2013-07-12 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Influence of the Agrc-Agra Complex in the Response Time of Staphylococcus Aureus Quorum Sensing
J.Bacteriol., 196, 2014
|
|
1JF7
 
 | | HUMAN PTP1B CATALYTIC DOMAIN COMPLEXED WITH PNU177836 | | Descriptor: | 5-(2-{2-[(TERT-BUTOXY-HYDROXY-METHYL)-AMINO]-1-HYDROXY-3-PHENYL-PROPYLAMINO}-3-HYDROXY-3-PENTYLAMINO-PROPYL)-2-CARBOXYMETHOXY-BENZOIC ACID, PROTEIN-TYROSINE PHOSPHATASE 1B | | Authors: | Larsen, S.D, Barf, T, Liljebris, C, May, P.D, Ogg, D, O'Sullivan, T.J, Palazuk, B.J, Schostarez, H.J, Stevens, F.C, Bleasdale, J.E. | | Deposit date: | 2001-06-20 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and biological activity of a novel class of small molecular weight peptidomimetic competitive inhibitors of protein tyrosine phosphatase 1B.
J.Med.Chem., 45, 2002
|
|
3I7T
 
 | |
3S3H
 
 | |
2PI7
 
 | |
4DCI
 
 | | Crystal structure of unknown funciton protein from Synechococcus sp. WH 8102 | | Descriptor: | SULFATE ION, uncharacterized protein | | Authors: | Chang, C, Marshall, N, Bearden, J, Palenik, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-17 | | Release date: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal structure of unknown function protein from Synechococcus sp. WH 8102
To be Published
|
|
2BSL
 
 | | Crystal structure of L. lactis dihydroorotate dehydrogense A in complex with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, ACETATE ION, DIHYDROOROTATE DEHYDROGENASE A, ... | | Authors: | Wolfe, A.E, Hansen, M, Gattis, S.G, Hu, Y.-C, Johansson, E, Arent, S, Larsen, S, Palfey, B.A. | | Deposit date: | 2005-05-23 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Interaction of Benzoate Pyrimidine Analogues with Class 1A Dihydroorotate Dehydrogenase from Lactococcus Lactis.
Biochemistry, 46, 2007
|
|
3Q83
 
 | |
2BX7
 
 | | Crystal structure of L. lactis dihydroorotate dehydrogense A in complex with 3,5-dihydroxybenzoate | | Descriptor: | 3,5-DIHYDROXYBENZOATE, ACETATE ION, DIHYDROOROTATE DEHYDROGENASE, ... | | Authors: | Wolfe, A.E, Hansen, M, Gattis, S.G, Hu, Y.-C, Johansson, E, Arent, S, Larsen, S, Palfey, B.A. | | Deposit date: | 2005-07-25 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Interaction of Benzoate Pyrimidine Analogues with Class 1A Dihydroorotate Dehydrogenase from Lactococcus Lactis.
Biochemistry, 46, 2007
|
|
3Q89
 
 | |
3Q8U
 
 | |
3Q8V
 
 | |
3Q86
 
 | |
