2AEO
 
 | | Crystal structure of cisplatinated bovine Cu,Zn superoxide dismutase | | Descriptor: | COPPER (II) ION, Cisplatin, Superoxide dismutase [Cu-Zn], ... | | Authors: | Calderone, V, Casini, A, Mangani, S, Messori, L, Orioli, P.L. | | Deposit date: | 2005-07-23 | | Release date: | 2006-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural investigation of cisplatin-protein interactions: selective platination of His19 in a cuprozinc superoxide dismutase.
Angew. Chem. Int. Ed. Engl., 45, 2006
|
|
5BU6
 
 | | Structure of BpsB deaceylase domain from Bordetella bronchiseptica | | Descriptor: | 1,2-ETHANEDIOL, BpsB (PgaB), Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase, ... | | Authors: | Little, D.J, Bamford, N.C, Howell, P.L. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | The Protein BpsB Is a Poly-beta-1,6-N-acetyl-d-glucosamine Deacetylase Required for Biofilm Formation in Bordetella bronchiseptica.
J.Biol.Chem., 290, 2015
|
|
1Z5P
 
 | | Crystal structure of MTA/AdoHcy nucleosidase with a ligand-free purine binding site | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Lee, J.E, Smith, G.D, Horvatin, C, Huang, D.J.T, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2005-03-18 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis
J.Mol.Biol., 352, 2005
|
|
1Z5O
 
 | | Crystal structure of MTA/AdoHcy nucleosidase Asp197Asn mutant complexed with 5'-methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Smith, G.D, Horvatin, C, Huang, D.J.T, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2005-03-18 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis
J.Mol.Biol., 352, 2005
|
|
5E7C
 
 | | Macromolecular diffractive imaging using imperfect crystals - Bragg data | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ayyer, K, Yefanov, O, Oberthuer, D, Roy-Chowdhury, S, Galli, L, Mariani, V, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Schaffner, A, Doerner, K, James, D, Kupitz, C, Metz, M, Nelson, G, Xavier, P.L, Beyerlein, K.R, Schmidt, M, Sarrou, I, Spence, J.C.H, Weierstall, U, White, T.A, Yang, J.-H, Zhao, Y, Liang, M, Aquila, A, Hunter, M.S, Robinson, J.S, Koglin, J.E, Boutet, S, Fromme, P, Barty, A, Chapman, H.N. | | Deposit date: | 2015-10-12 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Macromolecular diffractive imaging using imperfect crystals.
Nature, 530, 2016
|
|
1ZHG
 
 | | Crystal structure of Beta-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Plasmodium falciparum | | Descriptor: | beta hydroxyacyl-acyl carrier protein dehydratase | | Authors: | Swarnamukhi, P.L, Sharma, S.K, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2005-04-25 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of dimeric FabZ of Plasmodium falciparum reveals conformational switching to active hexamers by peptide flips
Febs Lett., 580, 2006
|
|
1Z5N
 
 | | Crystal structure of MTA/AdoHcy nucleosidase Glu12Gln mutant complexed with 5-methylthioribose and adenine | | Descriptor: | 5-S-methyl-5-thio-alpha-D-ribofuranose, ADENINE, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Smith, G.D, Horvatin, C, Huang, D.J.T, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2005-03-18 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis
J.Mol.Biol., 352, 2005
|
|
2AFP
 
 | | THE SOLUTION STRUCTURE OF TYPE II ANTIFREEZE PROTEIN REVEALS A NEW MEMBER OF THE LECTIN FAMILY | | Descriptor: | PROTEIN (SEA RAVEN TYPE II ANTIFREEZE PROTEIN) | | Authors: | Gronwald, W, Loewen, M.C, Lix, B, Daugulis, A.J, Sonnichsen, F.D, Davies, P.L, Sykes, B.D. | | Deposit date: | 1998-12-14 | | Release date: | 1998-12-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of type II antifreeze protein reveals a new member of the lectin family.
Biochemistry, 37, 1998
|
|
1Y6R
 
 | | Crystal structure of MTA/AdoHcy nucleosidase complexed with MT-ImmA. | | Descriptor: | (3S,4R)-2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-[(METHYLSULFANYL)METHYL]PYRROLIDINE-3,4-DIOL, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Singh, V, Evans, G.B, Tyler, P.C, Furneaux, R.H, Cornell, K.A, Riscoe, M.K, Schramm, V.L, Howell, P.L. | | Deposit date: | 2004-12-06 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
J.Biol.Chem., 280, 2005
|
|
4X7H
 
 | | Co-crystal Structure of PERK bound to N-{5-[(6,7-dimethoxyquinolin-4-yl)oxy]pyridin-2-yl}-1-methyl-3-oxo-2-phenyl-5-(pyridin-4-yl)-2,3-dihydro-1H-pyrazole-4-carboxamide inhibitor | | Descriptor: | Eukaryotic translation initiation factor 2-alpha kinase 3,Eukaryotic translation initiation factor 2-alpha kinase 3, N-{5-[(6,7-dimethoxyquinolin-4-yl)oxy]pyridin-2-yl}-1-methyl-3-oxo-2-phenyl-5-(pyridin-4-yl)-2,3-dihydro-1H-pyrazole-4-carboxamide, SULFATE ION | | Authors: | Shaffer, P.L, Bellon, S.F, Long, A.M, Chen, H. | | Deposit date: | 2014-12-09 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 1H-Pyrazol-3(2H)-ones as Potent and Selective Inhibitors of Protein Kinase R-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 58, 2015
|
|
4X7L
 
 | | Co-crystal Structure of PERK bound to 4-{2-amino-4-methyl-3-[2-(methylamino)-1,3-benzothiazol-6-yl]benzoyl}-1-methyl-2,5-diphenyl-1,2-dihydro-3H-pyrazol-3-one inhibitor | | Descriptor: | 4-{2-amino-4-methyl-3-[2-(methylamino)-1,3-benzothiazol-6-yl]benzoyl}-1-methyl-2,5-diphenyl-1,2-dihydro-3H-pyrazol-3-one, Eukaryotic translation initiation factor 2-alpha kinase 3,Eukaryotic translation initiation factor 2-alpha kinase 3, GLYCEROL, ... | | Authors: | Shaffer, P.L, Long, A.M, Chen, H. | | Deposit date: | 2014-12-09 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of 1H-Pyrazol-3(2H)-ones as Potent and Selective Inhibitors of Protein Kinase R-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 58, 2015
|
|
5E79
 
 | | Macromolecular diffractive imaging using imperfect crystals | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ayyer, K, Yefanov, O, Oberthur, D, Roy-Chowdhury, S, Galli, L, Mariani, V, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Schaffer, A, Dorner, K, James, D, Kupitz, C, Metz, M, Nelson, G, Xavier, P.L, Beyerlein, K.R, Schmidt, M, Sarrou, I, Spence, J.C.H, Weierstall, U, White, T.A, Yang, J.-H, Zhao, Y, Liang, M, Aquila, A, Hunter, M.S, Koglin, J.E, Boutet, S, Fromme, P, Barty, A, Chapman, H.N. | | Deposit date: | 2015-10-12 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Macromolecular diffractive imaging using imperfect crystals.
Nature, 530, 2016
|
|
6AME
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 M21A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
5EOY
 
 | | Pseudomonas aeruginosa SeMet-PilM bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type 4 fimbrial biogenesis protein PilM | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L.L, Howell, P.L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-27 | | Last modified: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
5EQ6
 
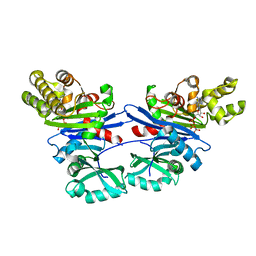 | | Pseudomonas aeruginosa PilM bound to AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Type 4 fimbrial biogenesis protein PilM | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L.L, Howell, P.L. | | Deposit date: | 2015-11-12 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
5EOX
 
 | | Pseudomonas aeruginosa PilM bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type 4 fimbrial biogenesis protein PilM | | Authors: | McCallum, M, Tammam, S, Robinson, H, Shah, M, Calmettes, C, Moraes, T, Burrows, L.L, Howell, P.L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
J.Biol.Chem., 291, 2016
|
|
5LLF
 
 | |
6VE4
 
 | | Pentadecameric PilQ from Pseudomonas aeruginosa | | Descriptor: | Fimbrial assembly protein PilQ | | Authors: | McCallum, M, Tammam, S, Rubinstein, J.L, Burrows, L.L, Howell, P.L. | | Deposit date: | 2019-12-28 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | CryoEM map of Pseudomonas aeruginosa PilQ enables structural characterization of TsaP.
Structure, 29, 2021
|
|
6BDT
 
 | |
6AU1
 
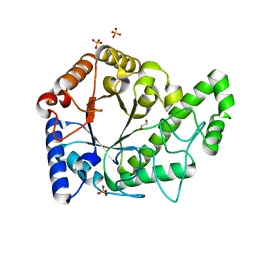 | | Structure of the PgaB (BpsB) glycoside hydrolase domain from Bordetella bronchiseptica | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative hemin storage protein, ... | | Authors: | Little, D.J, Bamford, N.C, Howell, P.L. | | Deposit date: | 2017-08-30 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | PgaB orthologues contain a glycoside hydrolase domain that cleaves deacetylated poly-beta (1,6)-N-acetylglucosamine and can disrupt bacterial biofilms.
PLoS Pathog., 14, 2018
|
|
5FNT
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-{4-Chloro-3-[(N-methylbenzenesulfonamido) methyl]phenyl}-3-(1-methyl-1H-1,2,3-benzotriazol-5-yl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FZJ
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | 2,6-DIMETHYL-4H-PYRANO[3,4-D][1,3]OXAZOL-4-ONE, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2016-03-14 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5MMX
 
 | | ABA RECEPTOR FROM CITRUS, CSPYL1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, CSPYL1_ABA | | Authors: | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L, Moreno-Alvero, M. | | Deposit date: | 2016-12-12 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.882 Å) | | Cite: | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
5MND
 
 | |
6BG8
 
 | |
