1TN7
 
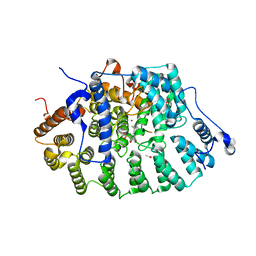 | | Protein Farnesyltransferase Complexed with a TC21 Peptide Substrate and a FPP Analog at 2.3A Resolution | | Descriptor: | ACETIC ACID, Fusion protein, Protein farnesyltransferase alpha subunit, ... | | Authors: | Reid, T.S, Terry, K.L, Casey, P.J, Beese, L.S. | | Deposit date: | 2004-06-11 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity.
J.Mol.Biol., 343, 2004
|
|
1TNU
 
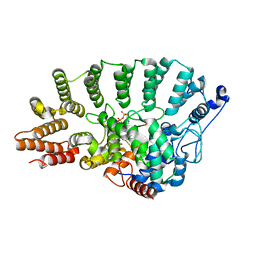 | | Rat Protein Geranylgeranyltransferase Type-I Complexed with a GGPP analog and a GCINCCKVL Peptide Derived from RhoB | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[METHYL-(5-GERANYL-4-METHYL-PENT-3-ENYL)-AMINO]-ETHYL-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Reid, T.S, Terry, K.L, Casey, P.J, Beese, L.S. | | Deposit date: | 2004-06-11 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity.
J.Mol.Biol., 343, 2004
|
|
1TNB
 
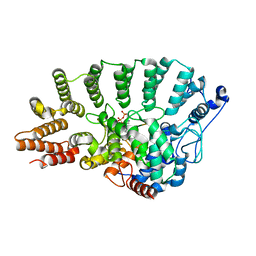 | | Rat Protein Geranylgeranyltransferase Type-I Complexed with a GGPP analog and a substrate KKSKTKCVIF Peptide Derived from TC21 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[METHYL-(5-GERANYL-4-METHYL-PENT-3-ENYL)-AMINO]-ETHYL-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Reid, T.S, Terry, K.L, Casey, P.J, Beese, L.S. | | Deposit date: | 2004-06-11 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity.
J.Mol.Biol., 343, 2004
|
|
2A9H
 
 | | NMR structural studies of a potassium channel / charybdotoxin complex | | Descriptor: | Voltage-gated potassium channel, charybdotoxin | | Authors: | Yu, L, Sun, C, Song, D, Shen, J, Xu, N, Gunasekera, A, Hajduk, P.J, Olejniczak, E.T. | | Deposit date: | 2005-07-11 | | Release date: | 2006-01-10 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structural studies of a potassium channel-charybdotoxin complex.
Biochemistry, 44, 2005
|
|
1TNO
 
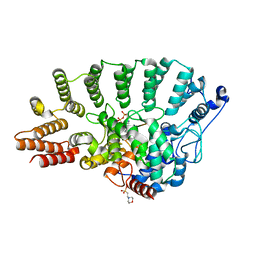 | | Rat Protein Geranylgeranyltransferase Type-I Complexed with a GGPP analog and a KKKSKTKCVIM Peptide Derived from K-Ras4B | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[METHYL-(5-GERANYL-4-METHYL-PENT-3-ENYL)-AMINO]-ETHYL-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Reid, T.S, Terry, K.L, Casey, P.J, Beese, L.S. | | Deposit date: | 2004-06-11 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity.
J.Mol.Biol., 343, 2004
|
|
3KZL
 
 | | Crystal structure of BA2930 mutant (H183G) in complex with AcCoA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-08 | | Release date: | 2009-12-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
1Z6O
 
 | | Crystal Structure of Trichoplusia ni secreted ferritin | | Descriptor: | CALCIUM ION, FE (III) ION, Ferritin heavy chain, ... | | Authors: | Hamburger, A.E, West Jr, A.P, Hamburger, Z.A, Hamburger, P, Bjorkman, P.J. | | Deposit date: | 2005-03-22 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of a secreted insect ferritin reveals a symmetrical arrangement of heavy and light chains.
J.Mol.Biol., 349, 2005
|
|
1DLP
 
 | | STRUCTURAL CHARACTERIZATION OF THE NATIVE FETUIN-BINDING PROTEIN SCILLA CAMPANULATA AGGLUTININ (SCAFET): A NOVEL TWO-DOMAIN LECTIN | | Descriptor: | LECTIN SCAFET PRECURSOR | | Authors: | Wright, L.M, Reynolds, C.D, Rizkallah, P.J, Allen, A.K, VanDamme, E.J.M, Donovan, M.J, Peumans, W.J. | | Deposit date: | 1999-12-11 | | Release date: | 2000-02-10 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural characterisation of the native fetuin-binding protein Scilla campanulata agglutinin: a novel two-domain lectin.
FEBS Lett., 468, 2000
|
|
1Z97
 
 | | Human Carbonic Anhydrase III: Structural and Kinetic Study of Catalysis and Proton Transfer. | | Descriptor: | Carbonic anhydrase III, ZINC ION | | Authors: | Duda, D.M, Tu, C, Fisher, S.Z, An, H, Yoshioka, C, Govindasamy, L, Laipis, P.J, Agbandje-McKenna, M, Silverman, D.N, McKenna, R. | | Deposit date: | 2005-03-31 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Carbonic Anhydrase III: Structural and Kinetic Study of Catalysis and Proton Transfer
Biochemistry, 44, 2005
|
|
1DOK
 
 | | MONOCYTE CHEMOATTRACTANT PROTEIN 1, P-FORM | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 1, SULFATE ION | | Authors: | Lubkowski, J, Bujacz, G, Boque, L, Wlodawer, A, Domaille, P.J, Handel, T.M. | | Deposit date: | 1996-11-27 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions.
Nat.Struct.Biol., 4, 1997
|
|
1T3P
 
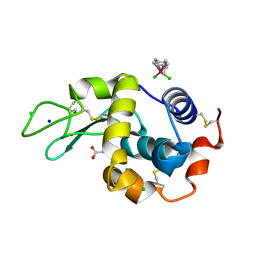 | | Half-sandwich arene ruthenium(II)-enzyme complex | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | McNae, I.W, Fishburne, K, Habtemariam, A, Hunter, T.M, Melchart, M, Wang, F, Walkinshaw, M.D, Sadler, P.J. | | Deposit date: | 2004-04-27 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Half-sandwich arene ruthenium(II)-enzyme complex
CHEM.COMMUN.(CAMB.), 16, 2004
|
|
1T1D
 
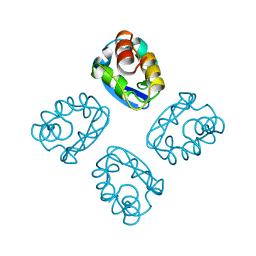 | |
2V8U
 
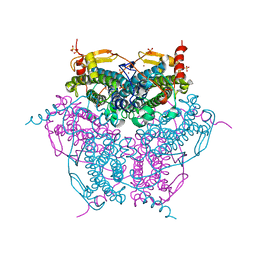 | | Atomic resolution structure of Mn catalase from Thermus Thermophilus | | Descriptor: | LITHIUM ION, MANGANESE (II) ION, MANGANESE-CONTAINING PSEUDOCATALASE, ... | | Authors: | Barynin, V.V, Antonyuk, S.V, Vaguine, A.A, Melik-Adamyan, W.R, Popov, A.N, Lamsin, V.S, Harrison, P.M, Artymiuk, P.J. | | Deposit date: | 2007-08-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Three-Dimentional Structure of the Enzyme Dimanganese Catalase from Thermus Thermophilus at 1 Angstrom Resolution
Crystallogr.Rep.(Transl. Kristallografiya), 45, 2000
|
|
1W4H
 
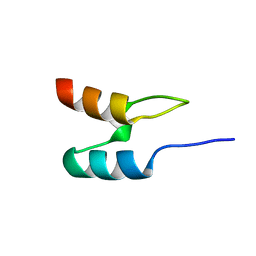 | | Peripheral-subunit from mesophilic, thermophilic and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1W7C
 
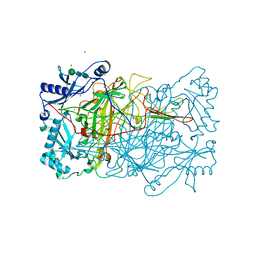 | | PPLO at 1.23 Angstroms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Duff, A.P, Cohen, A.E, Ellis, P.J, Guss, J.M. | | Deposit date: | 2004-09-01 | | Release date: | 2006-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | The 1.23 A Structure of Pichia Pastoris Lysyl Oxidase Reveals a Lysine-Lysine Cross-Link
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1W0E
 
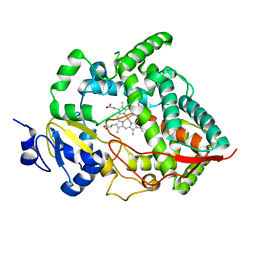 | | Crystal structure of human cytochrome P450 3A4 | | Descriptor: | CYTOCHROME P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Williams, P.A, Cosme, J, Vinkovic, D.M, Ward, A, Angove, H.C, Day, P.J, Vonrhein, C, Tickle, I.J, Jhoti, H. | | Deposit date: | 2004-06-03 | | Release date: | 2004-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Human Cytochrome P450 3A4 Bound to Metyrapone and Progesterone
Science, 305, 2004
|
|
1Z93
 
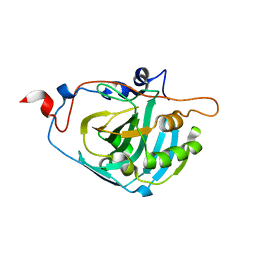 | | Human Carbonic Anhydrase III:Structural and Kinetic study of Catalysis and Proton Transfer. | | Descriptor: | Carbonic anhydrase III, ZINC ION | | Authors: | Duda, D.M, Tu, C, Fisher, S.Z, An, H, Yoshioka, C, Govindasamy, L, Laipis, P.J, Agbandje-McKenna, M, Silverman, D.N, McKenna, R. | | Deposit date: | 2005-03-31 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Carbonic Anhydrase III: Structural and Kinetic Study of Catalysis and Proton Transfer
Biochemistry, 44, 2005
|
|
1TC2
 
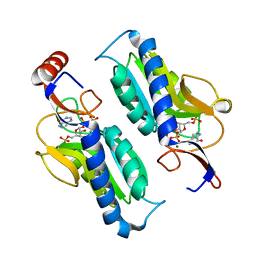 | | TERNARY SUBSTRATE COMPLEX OF THE HYPOXANTHINE PHOSPHORIBOSYLTRANSFERASE FROM TRYPANOSOMA CRUZI | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 7-HYDROXY-PYRAZOLO[4,3-D]PYRIMIDINE, MAGNESIUM ION, ... | | Authors: | Focia, P.J, Craig III, S.P, Eakin, A.E. | | Deposit date: | 1998-11-04 | | Release date: | 2000-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Approaching the transition state in the crystal structure of a phosphoribosyltransferase.
Biochemistry, 37, 1998
|
|
3LMJ
 
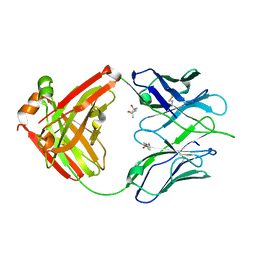 | | Structure of human anti HIV 21c Fab | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Heavy chain of anti HIV Fab from human 21c antibody, Light chain of anti HIV Fab from human 21c antibody | | Authors: | Diskin, R, Marcovecchio, P.M, Bjorkman, P.J. | | Deposit date: | 2010-01-30 | | Release date: | 2010-04-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1DQU
 
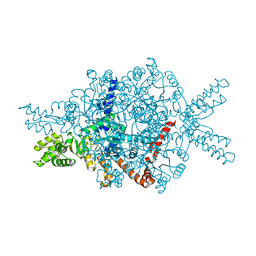 | | CRYSTAL STRUCTURE OF THE ISOCITRATE LYASE FROM ASPERGILLUS NIDULANS | | Descriptor: | ISOCITRATE LYASE | | Authors: | Britton, K.L, Langridge, S.J, Baker, P.J, Weeradechapon, K, Sedelnikova, S.E, De Lucas, J.R, Rice, D.W, Turner, G. | | Deposit date: | 2000-01-05 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure and active site location of isocitrate lyase from the fungus Aspergillus nidulans.
Structure Fold.Des., 8, 2000
|
|
3LQA
 
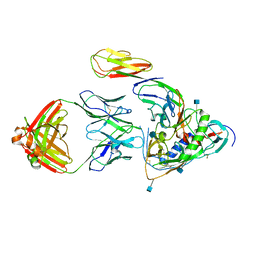 | | Crystal structure of clade C gp120 in complex with sCD4 and 21c Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, Heavy chain of anti HIV Fab from human 21c antibody, ... | | Authors: | Diskin, R, Marcovecchio, P.M, Bjorkman, P.J. | | Deposit date: | 2010-02-08 | | Release date: | 2010-04-07 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1XEE
 
 | | Solution structure of the Chemotaxis Inhibitory Protein of Staphylococcus aureus | | Descriptor: | chemotaxis-inhibiting protein CHIPS | | Authors: | Haas, P.J, de Haas, C.J, Poppelier, M.J, van Kessel, K.P, van Strijp, J.A, Dijkstra, K, Scheek, R.M, Fan, H, Kruijtzer, J.A, Liskamp, R.M, Kemmink, J. | | Deposit date: | 2004-09-10 | | Release date: | 2005-09-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of the C5a receptor-blocking domain of chemotaxis inhibitory protein of Staphylococcus aureus is related to a group of immune evasive molecules
J.Mol.Biol., 353, 2005
|
|
1VMP
 
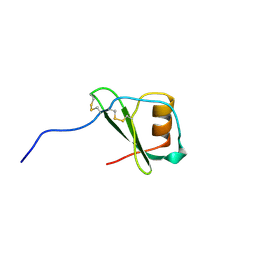 | | STRUCTURE OF THE ANTI-HIV CHEMOKINE VMIP-II | | Descriptor: | PROTEIN (ANTI-HIV CHEMOKINE MIP VII) | | Authors: | Liwang, A.C, Wang, Z.-X, Sun, Y, Peiper, S.C, Liwang, P.J. | | Deposit date: | 1999-03-25 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the anti-HIV chemokine vMIP-II.
Protein Sci., 8, 1999
|
|
1X9N
 
 | | Crystal Structure of Human DNA Ligase I bound to 5'-adenylated, nicked DNA | | Descriptor: | 5'-phosphorylated DNA, ADENOSINE MONOPHOSPHATE, DNA ligase I, ... | | Authors: | Pascal, J.M, O'Brien, P.J, Tomkinson, A.E, Ellenberger, T. | | Deposit date: | 2004-08-23 | | Release date: | 2004-11-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human DNA ligase I completely encircles and partially unwinds nicked DNA.
Nature, 432, 2004
|
|
1XD3
 
 | | Crystal structure of UCHL3-UbVME complex | | Descriptor: | MAGNESIUM ION, METHYL 4-AMINOBUTANOATE, UBC protein, ... | | Authors: | Misaghi, S, Galardy, P.J, Meester, W.J.N, Ovaa, H, Ploegh, H.L, Gaudet, R. | | Deposit date: | 2004-09-03 | | Release date: | 2004-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the Ubiquitin Hydrolase UCH-L3 Complexed with a Suicide Substrate
J.Biol.Chem., 280, 2005
|
|
