1N7X
 
 | | HUMAN SERUM TRANSFERRIN, N-LOBE Y45E MUTANT | | Descriptor: | CARBONATE ION, FE (III) ION, Serotransferrin | | Authors: | Adams, T.E, Mason, A.B, He, Q.Y, Halbrooks, P.J, Briggs, S.K, Smith, V.C, Macgillivray, R.T, Everse, S.J. | | Deposit date: | 2002-11-18 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | THE POSITION OF ARGININE 124 CONTROLS THE RATE OF IRON RELEASE FROM THE N-LOBE OF HUMAN SERUM TRANSFERRIN. A STRUCTURAL STUDY
J.Biol.Chem., 278, 2003
|
|
4HUS
 
 | | Crystal structure of streptogramin group A antibiotic acetyltransferase VatA from Staphylococcus aureus in complex with virginiamycin M1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stogios, P.J, Minasov, G, Evdokimova, E, Wawrzak, Z, Yim, V, Krishnamoorthy, M, Di Leo, R, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-03 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Potential for Reduction of Streptogramin A Resistance Revealed by Structural Analysis of Acetyltransferase VatA.
Antimicrob.Agents Chemother., 58, 2014
|
|
1NAF
 
 | |
5TIW
 
 | |
1N7W
 
 | | Crystal Structure of Human Serum Transferrin, N-Lobe L66W mutant | | Descriptor: | CARBONATE ION, FE (III) ION, Serotransferrin | | Authors: | Adams, T.E, Mason, A.B, He, Q.Y, Halbrooks, P.J, Briggs, S.K, Smith, V.C, MacGillivray, R.T, Everse, S.J. | | Deposit date: | 2002-11-18 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Position of Arginine 124 Controls the Rate of Iron Release from the N-lobe of Human Serum Transferrin. A Structural Study
J.Biol.Chem., 278, 2003
|
|
4I5C
 
 | | The Jak1 kinase domain in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-oxo-3-[(3R)-3-(pyrrolo[2,3-b][1,2,3]triazolo[4,5-d]pyridin-1(6H)-yl)piperidin-1-yl]propanenitrile, Tyrosine-protein kinase JAK1 | | Authors: | Fong, R, Lupardus, P.J. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel triazolo-pyrrolopyridines as inhibitors of Janus kinase 1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3FGS
 
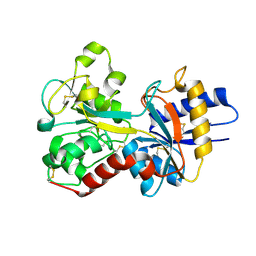 | |
7AQF
 
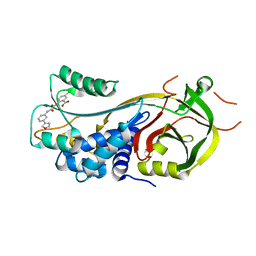 | |
5TVV
 
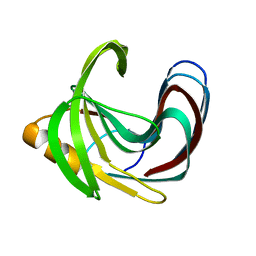 | | Computationally Designed Fentanyl Binder - Fen49* Apo | | Descriptor: | Endo-1,4-beta-xylanase A, POTASSIUM ION | | Authors: | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, A.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | Deposit date: | 2016-11-10 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
7AQG
 
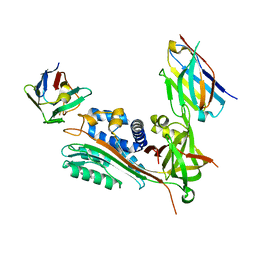 | | Crystal Structure of Small Molecule Inhibitor TM5484 Bound to Stabilized Active Plasminogen Activator Inhibitor-1 (PAI-1-W175F) | | Descriptor: | 5-Chloro-2-[[2-[3-(furan-3-yl)anilino]-2-oxoacetyl]amino]benzoic acid, Plasminogen activator inhibitor 1, VHH-2g-42 (Nb42), ... | | Authors: | Sillen, M, Strelkov, S.V, Declerck, P.J. | | Deposit date: | 2020-10-21 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Insight into the Two-Step Mechanism of PAI-1 Inhibition by Small Molecule TM5484.
Int J Mol Sci, 22, 2021
|
|
4HPL
 
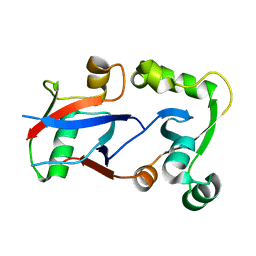 | | PCGF1 Ub fold (RAWUL)/BCOR PUFD Complex | | Descriptor: | BCL-6 corepressor, Polycomb group RING finger protein 1 | | Authors: | Junco, S.E, Wang, R, Gaipa, J, Taylor, A.B, Gearhart, M.D, Bardwell, V.J, Hart, P.J, Kim, C.A. | | Deposit date: | 2012-10-24 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Polycomb Group Protein PCGF1 in Complex with BCOR Reveals Basis for Binding Selectivity of PCGF Homologs.
Structure, 21, 2013
|
|
3CQP
 
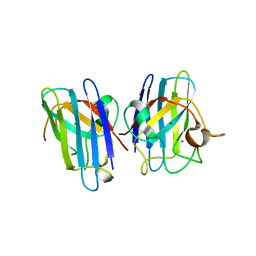 | | Human SOD1 G85R Variant, Structure I | | Descriptor: | COPPER (II) ION, MALONATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Cao, X, Antonyuk, S, Seetharaman, S.V, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Valentine, J.S, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2008-04-03 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the G85R Variant of SOD1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
7N35
 
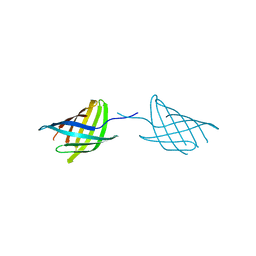 | |
7N34
 
 | |
3M52
 
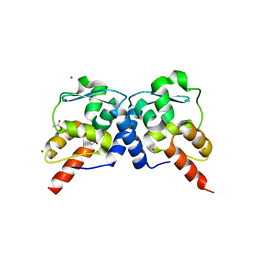 | | Crystal structure of the BTB domain from the Miz-1/ZBTB17 transcription regulator | | Descriptor: | ACETATE ION, ZINC ION, Zinc finger and BTB domain-containing protein 17 | | Authors: | Stogios, P.J, Cuesta-Seijo, J.A, Chen, L, Prive, G.G. | | Deposit date: | 2010-03-12 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Insights into Strand Exchange in BTB Domain Dimers from the Crystal Structures of FAZF and Miz1.
J.Mol.Biol., 400, 2010
|
|
5UEM
 
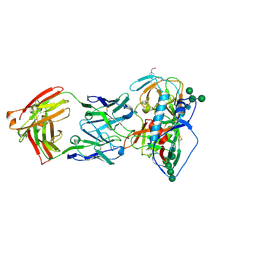 | | Crystal structure of 354NC37 Fab in complex with HIV-1 clade AE strain 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 354NC37 Fab Heavy Chain, ... | | Authors: | Sievers, S.A, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-01-02 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller.
Sci Transl Med, 9, 2017
|
|
5TP9
 
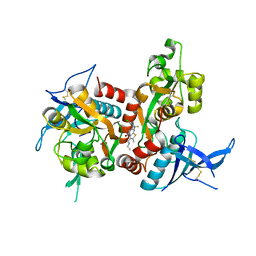 | | Structure of the human GluN1/GluN2A LBD in complex with compound 2 (GNE9178) | | Descriptor: | 7-{[5-chloro-3-(trifluoromethyl)-1H-pyrazol-1-yl]methyl}-N-ethyl-2-methyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-30 | | Last modified: | 2017-02-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | GluN2A-Selective Pyridopyrimidinone Series of NMDAR Positive Allosteric Modulators with an Improved in Vivo Profile.
ACS Med Chem Lett, 8, 2017
|
|
4RFQ
 
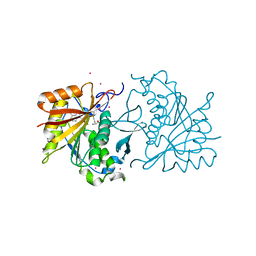 | | Human Methyltransferase-Like 18 | | Descriptor: | Histidine protein methyltransferase 1 homolog, S-ADENOSYLMETHIONINE, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Ravichandran, M, Li, Y, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Hong, B.S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-26 | | Release date: | 2014-10-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human Methyltransferase-Like 18
TO BE PUBLISHED
|
|
4I62
 
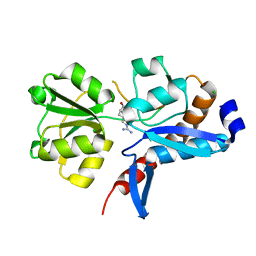 | | 1.05 Angstrom crystal structure of an amino acid ABC transporter substrate-binding protein AbpA from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine | | Descriptor: | ARGININE, Amino acid ABC transporter, periplasmic amino acid-binding protein, ... | | Authors: | Stogios, P.J, Kudritska, M, Wawrzak, Z, Minasov, G, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-29 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | 1.05 Angstrom crystal structure of an amino acid ABC transporter substrate-binding protein from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine
To be Published
|
|
3IFX
 
 | | Crystal structure of the Spin-labeled KcsA mutant V48R1 | | Descriptor: | POTASSIUM ION, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, TETRABUTYLAMMONIUM ION, ... | | Authors: | Cieslak, J.A, Focia, P.J, Gross, A. | | Deposit date: | 2009-07-26 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | EPR (3.56 Å), X-RAY DIFFRACTION | | Cite: | Electron Spin-Echo Envelope Modulation (ESEEM) Reveals Water and Phosphate Interactions with the KcsA Potassium Channel
Biochemistry, 49, 2010
|
|
5TTG
 
 | | Crystal structure of catalytic domain of GLP with MS012 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | DONG, A, ZENG, H, LIU, J, XIONG, Y, BABAULT, N, JIN, J, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, WU, H, BROWN, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-03 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase.
J. Med. Chem., 60, 2017
|
|
5U8J
 
 | | Crystal structure of a protein of unknown function ECL_02571 involved in membrane biogenesis from Enterobacter cloacae | | Descriptor: | CHLORIDE ION, UPF0502 protein BFJ73_07745 | | Authors: | Stogios, P.J, Skarina, T, Sandoval, J, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of a protein of unknown function ECL_02571 involved in membrane biogenesis from Enterobacter cloacae
To Be Published
|
|
1OIQ
 
 | | Imidazopyridines: a potent and selective class of Cyclin-dependent Kinase inhibitors identified through Structure-based hybridisation | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-[4-(2-METHYLIMIDAZO[1,2-A]PYRIDIN-3-YL)-2-PYRIMIDINYL]ACETAMIDE | | Authors: | Beattie, J.F, Breault, G.A, Byth, K.F, Culshaw, J.D, Ellston, R.P.A, Green, S, Minshull, C.A, Norman, R.A, Pauptit, R.A, Thomas, A.P, Jewsbury, P.J. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Imidazo[1,2-A]Pyridines: A Potent and Selective Class of Cyclin-Dependent Kinase Inhibitors Identified Through Structure-Based Hybridisation
Bioorg.Med.Chem.Lett., 13, 2003
|
|
4I3F
 
 | | Crystal structure of serine hydrolase CCSP0084 from the polyaromatic hydrocarbon (PAH)-degrading bacterium Cycloclasticus zankles | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Stogios, P.J, Xu, X, Dong, A, Cui, H, Alcaide, M, Tornes, J, Gertler, C, Yakimov, M.M, Golyshin, P.N, Ferrer, M, Savchenko, A. | | Deposit date: | 2012-11-26 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Single residues dictate the co-evolution of dual esterases: MCP hydrolases from the alpha / beta hydrolase family.
Biochem.J., 454, 2013
|
|
4QQ6
 
 | | Crystal Structure of tudor domain of SMN1 in complex with a small organic molecule | | Descriptor: | 4-methyl-2,3,4,5,6,7-hexahydrodicyclopenta[b,e]pyridin-8(1H)-imine, Survival motor neuron protein, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Iqbal, A, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-26 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
