1GJU
 
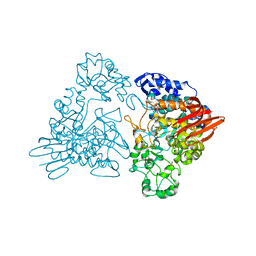 | | Maltosyltransferase from Thermotoga maritima | | Descriptor: | MALTODEXTRIN GLYCOSYLTRANSFERASE, PHOSPHATE ION | | Authors: | Roujeinikova, A, Raasch, C, Burke, J, Baker, P.J, Liebl, W, Rice, D.W. | | Deposit date: | 2001-08-02 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Maltosyltransferase and its Implications for the Molecular Basis of the Novel Transfer Specificity
J.Mol.Biol., 312, 2001
|
|
3C7N
 
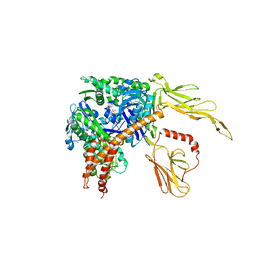 | | Structure of the Hsp110:Hsc70 Nucleotide Exchange Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CHLORIDE ION, ... | | Authors: | Schuermann, J.P, Jiang, J, Hart, P.J, Sousa, R. | | Deposit date: | 2008-02-07 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.115 Å) | | Cite: | Structure of the Hsp110:Hsc70 nucleotide exchange machine
Mol.Cell, 31, 2008
|
|
1ES9
 
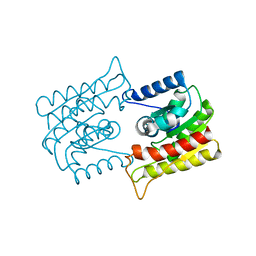 | | X-RAY CRYSTAL STRUCTURE OF R22K MUTANT OF THE MAMMALIAN BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASES (PAF-AH) | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE IB GAMMA SUBUNIT | | Authors: | McMullen, T.W.P, Li, J, Sheffield, P.J, Aoki, J, Martin, T.W, Arai, H, Inoue, K, Derewenda, Z.S. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The functional implications of the dimerization of the catalytic subunits of the mammalian brain platelet-activating factor acetylhydrolase (Ib).
Protein Eng., 13, 2000
|
|
1ERI
 
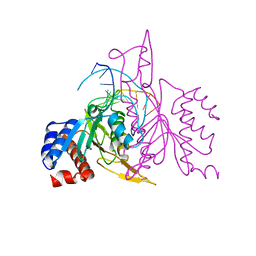 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE-DNA RECOGNITION COMPLEX: THE RECOGNITION NETWORK AND THE INTEGRATION OF RECOGNITION AND CLEAVAGE | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PROTEIN (ECO RI ENDONUCLEASE (E.C.3.1.21.4)) | | Authors: | Kim, Y, Grable, J.C, Love, R, Greene, P.J, Rosenberg, J.M. | | Deposit date: | 1994-05-18 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing.
Science, 249, 1990
|
|
1E5B
 
 | | Internal xylan binding domain from C. fimi Xyn10A, R262G mutant | | Descriptor: | XYLANASE D | | Authors: | Simpson, P.J, Hefang, X, Bolam, D.N, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2000-07-24 | | Release date: | 2001-05-25 | | Last modified: | 2018-10-24 | | Method: | SOLUTION NMR | | Cite: | The Structural Basis for the Ligand Specificity of Family 2 Carbohydrate Binding Nodules
J.Biol.Chem., 275, 2000
|
|
6VPN
 
 | |
1EQG
 
 | | THE 2.6 ANGSTROM MODEL OF OVINE COX-1 COMPLEXED WITH IBUPROFEN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IBUPROFEN, PROSTAGLANDIN H2 SYNTHASE-1, ... | | Authors: | Loll, P.J, Selinsky, B.S, Gupta, K, Sharkey, C.T. | | Deposit date: | 2000-04-04 | | Release date: | 2001-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural analysis of NSAID binding by prostaglandin H2 synthase: time-dependent and time-independent inhibitors elicit identical enzyme conformations.
Biochemistry, 40, 2001
|
|
6TMO
 
 | | Structure determination of an enhanced affinity TCR, a24b17, in complex with HLA-A*02:01 presenting a MART-1 peptide, EAAGIGILTV | | Descriptor: | 1,2-ETHANEDIOL, Alpha chain of engineered high affinity T-cell receptor, Beta chain of high affinity engineered T-cell receptor, ... | | Authors: | Rizkallah, P.J, Cole, D.K. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Rules Underpinning Enhanced Affinity Binding of Human T Cell Receptors Engineered for Immunotherapy.
Mol Ther Oncolytics, 18, 2020
|
|
1GU9
 
 | | Crystal Structure of Mycobacterium tuberculosis Alkylperoxidase AhpD | | Descriptor: | ALKYLHYDROPEROXIDASE D | | Authors: | Nunn, C.M, Djordjevic, S, Hillas, P.J, Nishida, C, Ortiz de Montellano, P.R. | | Deposit date: | 2002-01-24 | | Release date: | 2002-02-14 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Mycobacterium Tuberculosis Alkylhydroperoxidase Ahpd, a Potential Target for Antitubercular Drug Design
J.Biol.Chem., 277, 2002
|
|
3BLX
 
 | | Yeast Isocitrate Dehydrogenase (Apo Form) | | Descriptor: | Isocitrate dehydrogenase [NAD] subunit 1, Isocitrate dehydrogenase [NAD] subunit 2 | | Authors: | Taylor, A.B, Hu, G, Hart, P.J, McAlister-Henn, L. | | Deposit date: | 2007-12-11 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allosteric Motions in Structures of Yeast NAD+-specific Isocitrate Dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
3BUM
 
 | | Crystal structure of c-Cbl-TKB domain complexed with its binding motif in Sprouty2 | | Descriptor: | E3 ubiquitin-protein ligase CBL, Protein sprouty homolog 2 | | Authors: | Ng, C, Jackson, A.R, Buschdorf, P.J, Sun, Q, Guy, R.G, Sivaraman, J. | | Deposit date: | 2008-01-03 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates
Embo J., 27, 2008
|
|
1GJW
 
 | | Thermotoga maritima maltosyltransferase complex with maltose | | Descriptor: | MALTODEXTRIN GLYCOSYLTRANSFERASE, PHOSPHATE ION, alpha-D-glucopyranose, ... | | Authors: | Roujeinikova, A, Raasch, C, Burke, J, Baker, P.J, Liebl, W, Rice, D.W. | | Deposit date: | 2001-08-03 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Maltosyltransferase and its Implications for the Molecular Basis of the Novel Transfer Specificity
J.Mol.Biol., 312, 2001
|
|
6WON
 
 | | Crystal structure of acetoin dehydrogenase YohF from Salmonella typhimurium | | Descriptor: | CHLORIDE ION, SULFATE ION, YohF | | Authors: | Stogios, P.J, Skarina, T, Mesa, N, Endres, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-25 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of acetoin dehydrogenase YohF from Salmonella typhimurium
To Be Published
|
|
6WOP
 
 | | Crystal structure of gamma-aminobutyrate aminotransferase PuuE from Acinetobacter baumannii | | Descriptor: | 4-aminobutyrate transaminase, CHLORIDE ION, D(-)-TARTARIC ACID | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-25 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of gamma-aminobutyrate aminotransferase PuuE from Acinetobacter baumannii
To Be Published
|
|
6WT9
 
 | | Structure of STING-associated CdnE c-di-GMP synthase from Capnocytophaga granulosa | | Descriptor: | NTP_transf_2 domain-containing protein | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
3BLW
 
 | | Yeast Isocitrate Dehydrogenase with Citrate and AMP Bound in the Regulatory Subunits | | Descriptor: | ADENOSINE MONOPHOSPHATE, CITRATE ANION, Isocitrate dehydrogenase [NAD] subunit 1, ... | | Authors: | Taylor, A.B, Hu, G, Hart, P.J, McAlister-Henn, L. | | Deposit date: | 2007-12-11 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Allosteric Motions in Structures of Yeast NAD+-specific Isocitrate Dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
6WT6
 
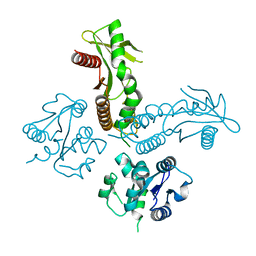 | | Structure of a metazoan TIR-STING receptor from C. gigas | | Descriptor: | Metazoan TIR-STING fusion | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
3C0C
 
 | |
1G0X
 
 | |
3BPR
 
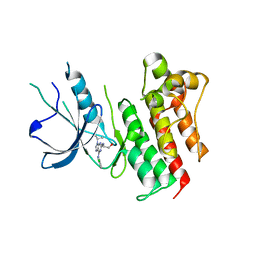 | | Crystal structure of catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor C52 | | Descriptor: | 2-(2-HYDROXYETHYLAMINO)-6-(3-CHLOROANILINO)-9-ISOPROPYLPURINE, CHLORIDE ION, Proto-oncogene tyrosine-protein kinase MER, ... | | Authors: | Walker, J.R, Huang, X, Finerty Jr, P.J, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-19 | | Release date: | 2008-01-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the inhibited states of the Mer receptor tyrosine kinase.
J.Struct.Biol., 165, 2009
|
|
1G8K
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ARSENITE OXIDASE FROM ALCALIGENES FAECALIS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ARSENITE OXIDASE, ... | | Authors: | Ellis, P.J, Conrads, T, Hille, R, Kuhn, P. | | Deposit date: | 2000-11-17 | | Release date: | 2000-12-13 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 A and 2.03 A.
Structure, 9, 2001
|
|
1H08
 
 | | CDK2 in complex with a disubstituted 2, 4-bis anilino pyrimidine CDK4 inhibitor | | Descriptor: | (2R)-1-{4-[(4-ANILINO-5-BROMOPYRIMIDIN-2-YL)AMINO]PHENOXY}-3-(DIMETHYLAMINO)PROPAN-2-OL, (2S)-1-{4-[(4-ANILINO-5-BROMOPYRIMIDIN-2-YL)AMINO]PHENOXY}-3-(DIMETHYLAMINO)PROPAN-2-OL, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Breault, G.A, Ellston, R.P.A, Green, S, James, S.R, Jewsbury, P.J, Midgley, C.J, Minshull, C.A, Pauptit, R.A, Tucker, J.A, Pease, J.E. | | Deposit date: | 2002-06-11 | | Release date: | 2003-07-11 | | Last modified: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclin-Dependent Kinase 4 Inhibitors as a Treatment for Cancer. Part 1: Identification and Optimisation of Substituted 4,6-Bis Anilino Pyrimidines
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1H01
 
 | | CDK2 in complex with a disubstituted 2, 4-bis anilino pyrimidine CDK4 inhibitor | | Descriptor: | (2R)-1-[4-({4-[(2,5-DICHLOROPHENYL)AMINO]PYRIMIDIN-2-YL}AMINO)PHENOXY]-3-(DIMETHYLAMINO)PROPAN-2-OL, (2S)-1-[4-({4-[(2,5-DICHLOROPHENYL)AMINO]PYRIMIDIN-2-YL}AMINO)PHENOXY]-3-(DIMETHYLAMINO)PROPAN-2-OL, CYCLIN-DEPENDENT KINASE 2, ... | | Authors: | Breault, G.A, Ellston, R.P.A, Green, S, James, S.R, Jewsbury, P.J, Midgley, C.J, Minshull, C.A, Pauptit, R.A, Tucker, J.A, Pease, J.E. | | Deposit date: | 2002-06-10 | | Release date: | 2003-07-11 | | Last modified: | 2011-10-19 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Cyclin-Dependent Kinase 4 Inhibitors as a Treatment for Cancer. Part 1: Identification and Optimisation of Substituted 4,6-Bis Anilino Pyrimidines
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1DON
 
 | |
1H07
 
 | | CDK2 in complex with a disubstituted 4, 6-bis anilino pyrimidine CDK4 inhibitor | | Descriptor: | ((2-BROMO-4-METHYLPHENYL){6-[(4-{[(2R)-3-(DIMETHYLAMINO)-2-HYDROXYPROPYL]OXY}PHENYL)AMINO]PYRIMIDIN-4-YL}AMINO)ACETONITRILE, ((2-BROMO-4-METHYLPHENYL){6-[(4-{[(2S)-3-(DIMETHYLAMINO)-2-HYDROXYPROPYL]OXY}PHENYL)AMINO]PYRIMIDIN-4-YL}AMINO)ACETONITRILE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Beattie, J.F, Breault, G.A, Ellston, R.P.A, Green, S, Jewsbury, P.J, Midgley, C.J, Naven, R.T, Minshull, C.A, Pauptit, R.A, Tucker, J.A, Pease, J.E. | | Deposit date: | 2002-06-11 | | Release date: | 2003-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cyclin-Dependent Kinase 4 Inhibitors as a Treatment for Cancer. Part 1: Identification and Optimisation of Substituted 4,6-Bis Anilino Pyrimidines
Bioorg.Med.Chem.Lett., 13, 2003
|
|
