6GUZ
 
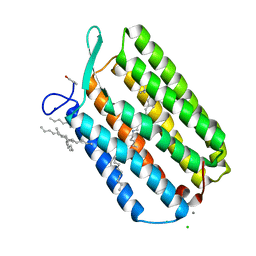 | | Ground state structure of Archaerhodopsin-3 obtained from LCP crystals using a thin-film sandwich at room temperature | | Descriptor: | Archaerhodopsin-3, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Moraes, I, Judge, P.J, Bada Juarez, J.F, Vinals, J, Axford, D, Watts, A. | | Deposit date: | 2018-06-19 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Room temperature structure of the Archaerhodopsin-3 obtained from LCP crystals using a thin-film sandwich
To Be Published
|
|
3EAM
 
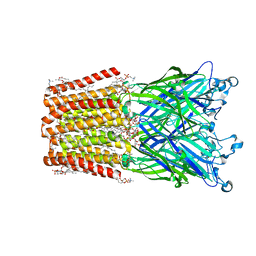 | | An open-pore structure of a bacterial pentameric ligand-gated ion channel | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Bocquet, N, Nury, H, Baaden, M, Le Poupon, C, Changeux, J.P, Delarue, M, Corringer, P.J. | | Deposit date: | 2008-08-26 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation.
Nature, 457, 2009
|
|
8SDC
 
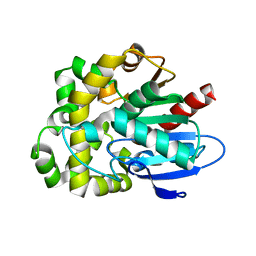 | | Crystal structure of fluoroacetate dehalogenase Daro3835 apoenzyme | | Descriptor: | Alpha/beta hydrolase fold protein, CHLORIDE ION | | Authors: | Stogios, P.J, Skarina, T, Khusnutdinova, A, Iakounine, A, Savchenko, A. | | Deposit date: | 2023-04-06 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural insights into hydrolytic defluorination of difluoroacetate by microbial fluoroacetate dehalogenases.
Febs J., 290, 2023
|
|
1ABZ
 
 | | ALPHA-T-ALPHA, A DE NOVO DESIGNED PEPTIDE, NMR, 23 STRUCTURES | | Descriptor: | ALPHA-T-ALPHA | | Authors: | Fezoui, Y, Connolly, P.J, Osterhout, J.J. | | Deposit date: | 1997-01-31 | | Release date: | 1998-02-04 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of alpha t alpha, a helical hairpin peptide of de novo design.
Protein Sci., 6, 1997
|
|
3BRB
 
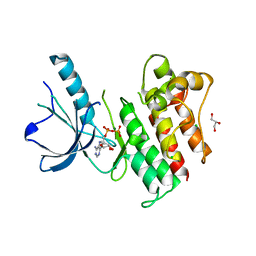 | | Crystal structure of catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Walker, J.R, Huang, X, Finerty Jr, P.J, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the inhibited states of the Mer receptor tyrosine kinase.
J.Struct.Biol., 165, 2009
|
|
6WT6
 
 | | Structure of a metazoan TIR-STING receptor from C. gigas | | Descriptor: | Metazoan TIR-STING fusion | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
3V03
 
 | | Crystal structure of Bovine Serum Albumin | | Descriptor: | ACETATE ION, CALCIUM ION, Serum albumin | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and immunologic characterization of bovine, horse, and rabbit serum albumins.
Mol.Immunol., 52, 2012
|
|
3C7N
 
 | | Structure of the Hsp110:Hsc70 Nucleotide Exchange Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CHLORIDE ION, ... | | Authors: | Schuermann, J.P, Jiang, J, Hart, P.J, Sousa, R. | | Deposit date: | 2008-02-07 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.115 Å) | | Cite: | Structure of the Hsp110:Hsc70 nucleotide exchange machine
Mol.Cell, 31, 2008
|
|
3UP1
 
 | | Crystal structure of the unliganded human interleukin-7 receptor extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | McElroy, C.A, Holland, P.J, Walsh, S.T.R. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-08 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural reorganization of the interleukin-7 signaling complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2VBD
 
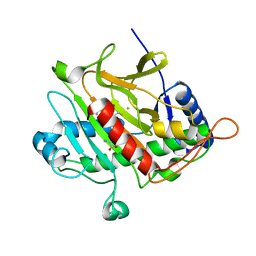 | | Isopenicillin N synthase with substrate analogue L,L,L-ACOMP (unexposed) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHETASE, N^6^-[(1R)-2-[(1R)-1-carboxy-2-(methylsulfanyl)ethoxy]-2-oxo-1-(sulfanylmethyl)ethyl]-6-oxo-L-lysine | | Authors: | Ge, W, Clifton, I.J, Adlington, R.M, Baldwin, J.E, Rutledge, P.J. | | Deposit date: | 2007-09-10 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of an Lll-Configured Depsipeptide Substrate Analogue Bound to Isopenicillin N Synthase.
Org.Biomol.Chem., 8, 2010
|
|
3V75
 
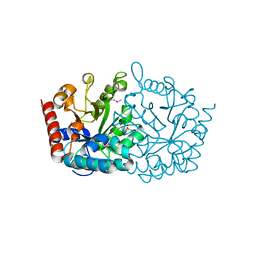 | | Crystal structure of putative orotidine 5'-phosphate decarboxylase from Streptomyces avermitilis ma-4680 | | Descriptor: | Orotidine 5'-phosphate decarboxylase | | Authors: | Stogios, P.J, Xu, X, Cui, H, Kudritska, M, Tan, K, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-12-20 | | Release date: | 2012-05-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of putative orotidine 5'-phosphate decarboxylase from Streptomyces avermitilis ma-4680
To be Published
|
|
3UV9
 
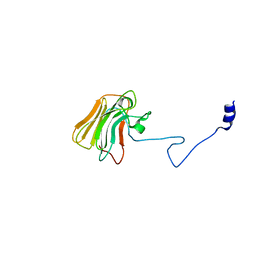 | | Structure of the rhesus monkey TRIM5alpha deltav1 PRYSPRY domain | | Descriptor: | Tripartite motif-containing protein 5 | | Authors: | Biris, N, Yang, Y, Taylor, A.B, Tomashevskii, A, Guo, M, Hart, P.J, Diaz-Griffero, F, Ivanov, D. | | Deposit date: | 2011-11-29 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Structure of the rhesus monkey TRIM5alpha PRYSPRY domain, the HIV capsid recognition module.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2VBB
 
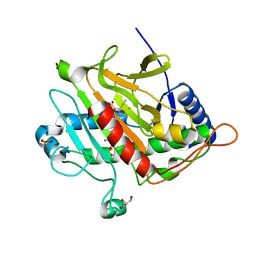 | | Isopenicillin N synthase with substrate analogue ACOMP (35minutes oxygen exposure) | | Descriptor: | FE (II) ION, GLYCEROL, ISOPENICILLIN N SYNTHETASE, ... | | Authors: | Ge, W, Clifton, I.J, Adlington, R.M, Baldwin, J.E, Rutledge, P.J. | | Deposit date: | 2007-09-07 | | Release date: | 2008-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Isopenicillin N Synthase Mediates Thiolate Oxidation to Sulfenate in a Depsipeptide Substrate Analogue: Implications for Oxygen Binding and a Link to Nitrile Hydratase?
J.Am.Chem.Soc., 130, 2008
|
|
6EH9
 
 | |
1D9X
 
 | | CRYSTAL STRUCTURE OF THE DNA REPAIR PROTEIN UVRB | | Descriptor: | EXCINUCLEASE UVRABC COMPONENT UVRB, ZINC ION | | Authors: | Theis, K, Chen, P.J, Skorvaga, M, Van Houten, B, Kisker, C. | | Deposit date: | 1999-10-30 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair.
EMBO J., 18, 1999
|
|
6WT5
 
 | | Structure of a bacterial STING receptor from Capnocytophaga granulosa | | Descriptor: | Bacterial STING | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
3V6O
 
 | | Leptin Receptor-antibody complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Carpenter, B, Hemsworth, G.R, Ross, R.J, Artymiuk, P.J. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the human obesity receptor leptin-binding domain reveals the mechanism of leptin antagonism by a monoclonal antibody.
Structure, 20, 2012
|
|
6WT8
 
 | | Structure of a STING-associated CdnE c-di-GMP synthase from Flavobacteriaceae sp. | | Descriptor: | STING-associated CdnE c-di-GMP synthase | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
2XL4
 
 | | LntA, a virulence factor from Listeria monocytogenes | | Descriptor: | GLYCEROL, Listeria nuclear targeted protein A | | Authors: | Lebreton, A, Job, V, Tham, T.N, Camejo, A, Mattei, P.J, Regnault, B, Cabanes, D, Dessen, A, Cossart, P, Bierne, H. | | Deposit date: | 2010-07-19 | | Release date: | 2011-02-02 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response.
Science, 331, 2011
|
|
4A69
 
 | | Structure of HDAC3 bound to corepressor and inositol tetraphosphate | | Descriptor: | ACETATE ION, D-MYO INOSITOL 1,4,5,6 TETRAKISPHOSPHATE, GLYCEROL, ... | | Authors: | Watson, P.J, Fairall, L, Santos, G.M, Schwabe, J.W.R. | | Deposit date: | 2011-11-01 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of Hdac3 Bound to Co-Repressor and Inositol Tetraphosphate.
Nature, 481, 2012
|
|
6GWP
 
 | | Crystal Structure of Stabilized Active Plasminogen Activator Inhibitor-1 (PAI-1-stab) in Complex with Two Inhibitory Nanobodies (VHH-2g-42, VHH-2w-64) | | Descriptor: | Plasminogen Activator Inhibitor-1, VHH-2g-42, VHH-2w-64 | | Authors: | Sillen, M, Weeks, S.D, Strelkov, S.V, Declerck, P.J. | | Deposit date: | 2018-06-25 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Molecular mechanism of two nanobodies that inhibit PAI-1 activity reveals a modulation at distinct stages of the PAI-1/plasminogen activator interaction.
J.Thromb.Haemost., 18, 2020
|
|
2XTD
 
 | | Structure of the TBL1 tetramerisation domain | | Descriptor: | TBL1 F-BOX-LIKE/WD REPEAT-CONTAINING PROTEIN TBL1X | | Authors: | Oberoi, J, Fairall, L, Watson, P.J, Greenwood, J.A, Schwabe, J.W.R. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for the Assembly of the Smrt/Ncor Core Transcriptional Repression Machinery.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2VWP
 
 | | Haloferax mediterranei glucose dehydrogenase in complex with NADPH and Zn. | | Descriptor: | GLUCOSE DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Baker, P.J, Britton, K.L, Fisher, M, Esclapez, J, Pire, C, Bonete, M.J, Ferrer, J, Rice, D.W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Active site dynamics in the zinc-dependent medium chain alcohol dehydrogenase superfamily.
Proc. Natl. Acad. Sci. U.S.A., 106, 2009
|
|
6EH7
 
 | |
1C0Q
 
 | | COMPLEX OF VANCOMYCIN WITH 2-ACETOXY-D-PROPANOIC ACID | | Descriptor: | CHLORIDE ION, LACTIC ACID, VANCOMYCIN, ... | | Authors: | Loll, P.J, Kaplan, J, Selinsky, B, Axelsen, P.H. | | Deposit date: | 1999-07-20 | | Release date: | 1999-07-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Vancomycin binding to low-affinity ligands: delineating a minimum set of interactions necessary for high-affinity binding.
J.Med.Chem., 42, 1999
|
|
