7Y4B
 
 | | Crystal structure of DUSP10 mutant_D59A | | Descriptor: | Dual specificity protein phosphatase 10 | | Authors: | Hu, I.C, Lyu, P.C. | | Deposit date: | 2022-06-14 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of DUSP10 mutant_D59A
To Be Published
|
|
7Y4D
 
 | | Crystal structure of DUSP10 mutant_S95A | | Descriptor: | Dual specificity protein phosphatase 10 | | Authors: | Hu, I.C, Lyu, P.C. | | Deposit date: | 2022-06-14 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of DUSP10 mutant_S95A
To Be Published
|
|
7Y4C
 
 | | Crystal structure of DUSP10 | | Descriptor: | Dual specificity protein phosphatase 10 | | Authors: | Hu, I.C, Lyu, P.C. | | Deposit date: | 2022-06-14 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of DUSP10
To Be Published
|
|
6BC0
 
 | |
2GHK
 
 | | Conformational mobility in the active site of a heme peroxidase | | Descriptor: | CYANIDE ION, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2006-03-27 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
5C3X
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I72D/N100E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Skerritt, L.A, Bell-Upp, P.C, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS I72D/N100E at cryogenic temperature
To be Published
|
|
7YM1
 
 | | Structure of SsbA protein in complex with the anticancer drug 5-fluorouracil | | Descriptor: | 5-FLUOROURACIL, GLYCEROL, Single-stranded DNA-binding protein | | Authors: | Huang, Y.H, Yang, P.C, Chiang, W.Y, Lin, E.S, Huang, C.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal Structure of DNA Replication Protein SsbA Complexed with the Anticancer Drug 5-Fluorouracil.
Int J Mol Sci, 24, 2023
|
|
6BC1
 
 | |
6BCA
 
 | |
2GL1
 
 | | NMR solution structure of Vigna radiata Defensin 2 (VrD2) | | Descriptor: | PDF1 | | Authors: | Lin, K.F, Lee, T.R, Tsai, P.H, Hsu, M.P, Chen, C.S, Lyu, P.C. | | Deposit date: | 2006-04-04 | | Release date: | 2007-04-03 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure-based protein engineering for alpha-amylase inhibitory activity of plant defensin.
Proteins, 68, 2007
|
|
7ZDQ
 
 | | Cryo-EM structure of Human ACE2 bound to a high-affinity SARS CoV-2 mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Bate, N, Savva, C.G, Moody, P.C.E, Brown, E.A, Schwabe, W.R, Brindle, N.P.J, Ball, J.K, Sale, J.E. | | Deposit date: | 2022-03-29 | | Release date: | 2022-05-18 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | In vitro evolution predicts emerging SARS-CoV-2 mutations with high affinity for ACE2 and cross-species binding.
Plos Pathog., 18, 2022
|
|
6B9B
 
 | |
5C5L
 
 | |
5CC4
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS T62E/V66E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Skerritt, L.A, Bell-Upp, P.C, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-07-01 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS T62E/V66E at cryogenic temperature
To be Published
|
|
5C5M
 
 | |
5CGK
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66D/N100E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Skerritt, L.A, Bell-Upp, P.C, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-07-09 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66D/N100E at cryogenic temperature
To be Published
|
|
5JQR
 
 | | The Structure of Ascorbate Peroxidase Compound II formed by reaction with m-CPBA | | Descriptor: | Ascorbate peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kwon, H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2016-05-05 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Direct visualization of a Fe(IV)-OH intermediate in a heme enzyme.
Nat Commun, 7, 2016
|
|
1XMJ
 
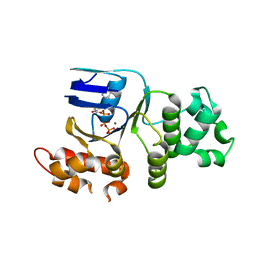 | | Crystal structure of human deltaF508 human NBD1 domain with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Lewis, H.A, Zhao, X, Wang, C, Sauder, J.M, Rooney, I, Noland, B.W, Lorimer, D, Kearins, M.C, Conners, K, Condon, B, Maloney, P.C, Guggino, W.B, Hunt, J.F, Emtage, S, Structural GenomiX | | Deposit date: | 2004-10-02 | | Release date: | 2004-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Impact of the delta-F508 Mutation in First Nucleotide-binding Domain of Human Cystic Fibrosis Transmembrane Conductance Regulator on Domain Folding and Structure
J.Biol.Chem., 280, 2005
|
|
3J7Y
 
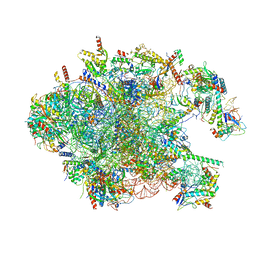 | | Structure of the large ribosomal subunit from human mitochondria | | Descriptor: | 16S rRNA, ADENOSINE MONOPHOSPHATE, CRIF1, ... | | Authors: | Brown, A, Amunts, A, Bai, X.C, Sugimoto, Y, Edwards, P.C, Murshudov, G, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the large ribosomal subunit from human mitochondria.
Science, 346, 2014
|
|
6CFQ
 
 | |
5CR6
 
 | | Structure of pneumolysin at 1.98 A resolution | | Descriptor: | Pneumolysin | | Authors: | Marshall, J.E, Faraj, B.H.A, Gingras, A.R, Lonnen, R, Sheikh, M.A, El-Mezgueldi, M, Moody, P.C.E, Andrew, P.W, Wallis, R. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Crystal Structure of Pneumolysin at 2.0 angstrom Resolution Reveals the Molecular Packing of the Pre-pore Complex.
Sci Rep, 5, 2015
|
|
1VRP
 
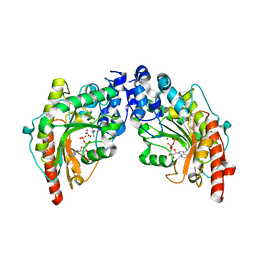 | | The 2.1 Structure of T. californica Creatine Kinase Complexed with the Transition-State Analogue Complex, ADP-Mg 2+ /NO3-/Creatine | | Descriptor: | (DIAMINOMETHYL-METHYL-AMINO)-ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, Creatine Kinase, ... | | Authors: | Lahiri, S.D, Wang, P.F, Babbitt, P.C, McLeish, M.J, Kenyon, G.L, Allen, K.N. | | Deposit date: | 2005-04-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1 A Structure of Torpedo californica Creatine Kinase Complexed with the ADP-Mg(2+)-NO3(-)-Creatine Transition-State Analogue Complex
Biochemistry, 41, 2002
|
|
1VYP
 
 | | Structure of pentaerythritol tetranitrate reductase W102F mutant and complexed with picric acid | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE, PICRIC ACID | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2004-05-04 | | Release date: | 2004-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Atomic Resolution Structures and Solution Behavior of Enzyme-Substrate Complexes of Enterobacter Cloacae Pb2 Pentaerythritol Tetranitrate Reductase: Multiple Conformational States and Implications for the Mechanism of Nitroaromatic Explosive Degradation
J.Biol.Chem., 279, 2004
|
|
6CEK
 
 | |
6CDQ
 
 | |
