2XKR
 
 | | Crystal Structure of Mycobacterium tuberculosis CYP142: A novel cholesterol oxidase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 142, TETRAETHYLENE GLYCOL | | Authors: | Driscoll, M, McLean, K.J, Levy, C.W, Lafite, P, Mast, N, Pikuleva, I.A, Rigby, S.E.J, Leys, D, Munro, A.W. | | Deposit date: | 2010-07-12 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and Biochemical Characterization of Mycobacterium Tuberculosis Cyp142: Evidence for Multiple Cholesterol 27-Hydroxylase Activities in a Human Pathogen.
J.Biol.Chem., 285, 2010
|
|
3PT2
 
 | | Structure of a viral OTU domain protease bound to Ubiquitin | | Descriptor: | 1.7.6 3-bromanylpropan-1-amine, ACETATE ION, RNA polymerase, ... | | Authors: | James, T.W, Bacik, J.P, Frias-Staheli, N, Garcia-Sastre, A, Mark, B.L. | | Deposit date: | 2010-12-02 | | Release date: | 2011-01-19 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PYP
 
 | | PHOTOACTIVE YELLOW PROTEIN, CRYOTRAPPED EARLY LIGHT CYCLE INTERMEDIATE | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Genick, U.K, Soltis, S.M, Kuhn, P, Canestrelli, I.L, Getzoff, E.D. | | Deposit date: | 1998-07-28 | | Release date: | 1999-06-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Structure at 0.85 A resolution of an early protein photocycle intermediate.
Nature, 392, 1998
|
|
2XT6
 
 | | Crystal structure of Mycobacterium smegmatis alpha-ketoglutarate decarboxylase homodimer (orthorhombic form) | | Descriptor: | 2-OXOGLUTARATE DECARBOXYLASE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2010-10-05 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
5YW4
 
 | | Structure-Guided Engineering of Reductase: Efficient Attenuating Substrate Inhibition in Asymmetric Catalysis | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Protein induced by osmotic stress | | Authors: | Shang, Y.P, Chen, Q, Li, A.T, Yu, H.L, Xu, J.H. | | Deposit date: | 2017-11-28 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Attenuated substrate inhibition of a haloketone reductase via structure-guided loop engineering.
J.Biotechnol., 308, 2020
|
|
5ZHP
 
 | | M3 muscarinic acetylcholine receptor in complex with a selective antagonist | | Descriptor: | (1R,2R,4S,5S,7s)-7-({[4-fluoro-2-(thiophen-2-yl)phenyl]carbamoyl}oxy)-9,9-dimethyl-3-oxa-9-azatricyclo[3.3.1.0~2,4~]nonan-9-ium, CITRIC ACID, HEXAETHYLENE GLYCOL, ... | | Authors: | Liu, H, Hofmann, J, Fish, I, Schaake, B, Eitel, K, Bartuschat, A, Kaindl, J, Rampp, H, Banerjee, A, Hubner, H, Clark, M.J, Vincent, S.G, Fisher, J, Heinrich, M, Hirata, K, Liu, X, Sunahara, R.K, Shoichet, B.K, Kobilka, B.K, Gmeiner, P. | | Deposit date: | 2018-03-13 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-guided development of selective M3 muscarinic acetylcholine receptor antagonists
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YYA
 
 | | Crystal structure of Tandem Tudor Domain of human UHRF1 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, SULFATE ION | | Authors: | Kori, S, Defossez, P.A, Arita, K. | | Deposit date: | 2017-12-08 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the UHRF1 Tandem Tudor Domain Bound to a Methylated Non-histone Protein, LIG1, Reveals Rules for Binding and Regulation.
Structure, 27, 2019
|
|
5XK4
 
 | | Retracted state of S65-phosphorylated ubiquitin | | Descriptor: | Polyubiquitin-B | | Authors: | Dong, X, Gong, Z, Qin, L.Y, Ran, M.L, Zhang, C.L, Liu, K, Liu, Z, Zhang, W.P, Tang, C. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2Y61
 
 | | Crystal structure of Leishmanial E65Q-TIM complexed with S-Glycidol phosphate | | Descriptor: | GLYCEROL, SN-GLYCEROL-1-PHOSPHATE, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Venkatesan, R, Alahuhta, M, Pihko, P.M, Wierenga, R.K. | | Deposit date: | 2011-01-19 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | High resolution crystal structures of triosephosphate isomerase complexed with its suicide inhibitors: the conformational flexibility of the catalytic glutamate in its closed, liganded active site.
Protein Sci., 20, 2011
|
|
2XV4
 
 | | Structure of Human RPC62 (partial) | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC3, PHOSPHATE ION | | Authors: | Lefevre, S, Legrand, P, Fribourg, S. | | Deposit date: | 2010-10-22 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-Function Analysis of Hrpc62 Provides Insights Into RNA Polymerase III Transcription
Nat.Struct.Mol.Biol., 18, 2011
|
|
5XK5
 
 | | Relaxed state of S65-phosphorylated ubiquitin | | Descriptor: | Polyubiquitin-B | | Authors: | Xu, D, Zhou, G, Qin, L.Y, Ran, M.L, Zhang, C.L, Liu, K, Liu, Z, Zhang, W.P, Tang, C. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2XU7
 
 | | Structural basis for RbAp48 binding to FOG-1 | | Descriptor: | HISTONE-BINDING PROTEIN RBBP4, TETRAETHYLENE GLYCOL, ZINC FINGER PROTEIN ZFPM1 | | Authors: | Lejon, S, Thong, S.Y, Murthy, A, Blobel, G.A, Mackay, J.P, Murzina, N.V, Laue, E.D. | | Deposit date: | 2010-10-15 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights Into Association of the Nurd Complex with Fog-1 from the Crystal Structure of an Rbap48-Fog- 1 Complex.
J.Biol.Chem., 286, 2011
|
|
2XWX
 
 | | Vibrio cholerae colonization factor GbpA crystal structure | | Descriptor: | GLCNAC-BINDING PROTEIN A | | Authors: | Wong, E, Vaaje-Kolstad, G, Ghosh, A, Guerrero, R.H, Konarev, P.V, Ibrahim, A.F.M, Svergun, D.I, Eijsink, V.G.H, Chatterjee, N.S, van Aalten, D.M.F. | | Deposit date: | 2010-11-06 | | Release date: | 2011-11-16 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Vibrio Cholerae Colonization Factor Gbpa Possesses a Modular Structure that Governs Binding to Different Host Surfaces.
Plos Pathog., 8, 2012
|
|
3N4P
 
 | | Human cytomegalovirus terminase nuclease domain | | Descriptor: | MAGNESIUM ION, Terminase subunit UL89 protein | | Authors: | Nadal, M, Mas, P.J, Blanco, A.G, Arnan, C, Sola, M, Hart, D.J, Coll, M. | | Deposit date: | 2010-05-22 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3N5G
 
 | | Crystal Structure of histidine-tagged human thymidylate synthase | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Pozzi, C, Cardinale, D, Guaitoli, G, Tondi, D, Luciani, R, Myllykallio, H, Ferrari, S, Costi, M.P, Mangani, S. | | Deposit date: | 2010-05-25 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Protein-protein interface-binding peptides inhibit the cancer therapy target human thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2XU6
 
 | | MDV1 coiled coil domain | | Descriptor: | MDV1 COILED COIL | | Authors: | Koirala, S, Bui, H.T, Schubert, H.L, Eckert, D.M, Hill, C.P, Kay, M.S, Shaw, J.M. | | Deposit date: | 2010-10-14 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Architecture of a Dynamin Adaptor: Implications for Assembly of Mitochondrial Fission Complexes
J.Cell Biol., 191, 2010
|
|
2XV5
 
 | | Human lamin A coil 2B fragment | | Descriptor: | LAMIN-A/C | | Authors: | Kapinos, L.E, Burkhard, P, Aebi, U, Herrmann, H, Strelkov, S.V. | | Deposit date: | 2010-10-22 | | Release date: | 2011-03-09 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Simultaneous Formation of Right- and Left-Handed Anti-Parallel Coiled-Coil Interfaces by a Coil2 Fragment of Human Lamin A.
J.Mol.Biol., 408, 2011
|
|
5XYJ
 
 | |
2YX4
 
 | | Crystal Structure of T134A of ST1022 from Sulfolobus tokodaii | | Descriptor: | 150aa long hypothetical transcriptional regulator, GLUTAMINE, MAGNESIUM ION | | Authors: | Kumarevel, T.S, Karthe, P, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-24 | | Release date: | 2008-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding
Nucleic Acids Res., 36, 2008
|
|
2YN2
 
 | | Huf protein - paralogue of the tau55 histidine phosphatase domain | | Descriptor: | FORMIC ACID, UNCHARACTERIZED PROTEIN YNL108C | | Authors: | Taylor, N.M.I, Glatt, S, Hennrich, M, von Scheven, G, Grotsch, H, Fernandez-Tornero, C, Rybin, V, Gavin, A.C, Kolb, P, Muller, C.W. | | Deposit date: | 2012-10-11 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Functional Characterization of a Phosphatase Domain within Yeast General Transcription Factor Tfiiic.
J.Biol.Chem., 288, 2013
|
|
2Y6B
 
 | | Ascorbate Peroxidase R38K mutant | | Descriptor: | ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Metcalfe, C.L, Efimov, I, Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2011-01-20 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Proton Delivery to Ferryl Heme in a Heme Peroxidase: Enzymatic Use of the Grotthuss Mechanism.
J.Am.Chem.Soc., 133, 2011
|
|
5XYB
 
 | |
7Z44
 
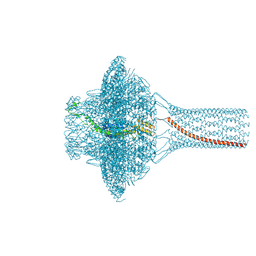 | | Portal of bacteriophage SU10 | | Descriptor: | Portal protein | | Authors: | Siborova, M, Fuzik, T, Prochazkova, M, Novacek, J, Plevka, P. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Tail proteins of phage SU10 reorganize into the nozzle for genome delivery.
Nat Commun, 13, 2022
|
|
2Y8A
 
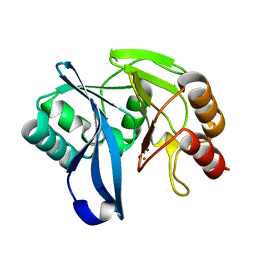 | | VIM-7 with Oxidised. Structural and computational investigations of VIM-7: Insights into the substrate specificity of VIM metallo-beta- lactamases | | Descriptor: | MAGNESIUM ION, METALLO-B-LACTAMASE, UNKNOWN ATOM OR ION, ... | | Authors: | Saradhi, P, Leiros, H.-K.S, Ahmad, R, Spencer, J, Leiros, I, Walsh, T.R, Sundsfjord, A, Samuelsen, O. | | Deposit date: | 2011-02-03 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural and Computational Investigations of Vim- 7: Insights Into the Substrate Specificity of Vim Metallo-Beta-Lactamases
J.Mol.Biol., 411, 2011
|
|
3N9Q
 
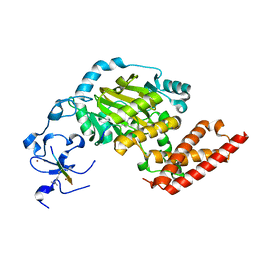 | | ceKDM7A from C.elegans, complex with H3K4me3 peptide, H3K27me2 peptide and NOG | | Descriptor: | FE (II) ION, Histone H3 peptide, N-OXALYLGLYCINE, ... | | Authors: | Yang, Y, Hu, L, Wang, P, Hou, H, Chen, C.D, Xu, Y. | | Deposit date: | 2010-05-31 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans
Cell Res., 20, 2010
|
|
