5DS9
 
 | |
4O9S
 
 | | Crystal structure of Retinol-Binding Protein 4 (RBP4)in complex with a non-retinoid ligand | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(12),2(6),8,10-tetraen-12-yl)piperazin-1-yl]-2-[2-(trifluoromethyl)phenyl]ethanone, CHLORIDE ION, ... | | Authors: | Wang, Z, Johnstone, S, Walker, N.P. | | Deposit date: | 2014-01-02 | | Release date: | 2014-07-02 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-assisted discovery of the first non-retinoid ligands for Retinol-Binding Protein 4.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
6RSD
 
 | |
6H8E
 
 | | Truncated derivative of the C-terminal domain of the TssA component of the type VI secretion system from Burkholderia cenocepacia | | Descriptor: | Type VI secretion protein ImpA | | Authors: | Dix, S.R, Owen, H.J, Sun, R, Ahmad, A, Shastri, S, Spiewak, H.L, Mosby, D.J, Harris, M.J, Batters, S.L, Brooker, T.A, Tzokov, S.B, Sedelnikova, S.E, Baker, P.J, Bullough, P.A, Rice, D.W, Thomas, M.S. | | Deposit date: | 2018-08-02 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into the function of type VI secretion system TssA subunits.
Nat Commun, 9, 2018
|
|
8QN3
 
 | | OPR3 wildtype in complex with NADH4 | | Descriptor: | 1,4,5,6-Tetrahydronicotinamide adenine dinucleotide, 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Bijelic, A, Macheroux, P, Keschbaumer, B. | | Deposit date: | 2023-09-25 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Loop 6 and the beta-hairpin flap are structural hotspots that determine cofactor specificity in the FMN-dependent family of ene-reductases.
Febs J., 291, 2024
|
|
8QOH
 
 | | Crystal structure of the kinetoplastid kinetochore protein KKT14 C-terminal domain from Apiculatamorpha spiralis | | Descriptor: | kinetochore protein KKT14 | | Authors: | Carter, W, Ballmer, D, Ishii, M, Ludzia, P, Akiyoshi, B. | | Deposit date: | 2023-09-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetoplastid kinetochore proteins KKT14-KKT15 are divergent Bub1/BubR1-Bub3 proteins.
Open Biology, 14, 2024
|
|
1QAD
 
 | | Crystal Structure of the C-Terminal SH2 Domain of the P85 alpha Regulatory Subunit of Phosphoinositide 3-Kinase: An SH2 domain mimicking its own substrate | | Descriptor: | PI3-KINASE P85 ALPHA SUBUNIT | | Authors: | Hoedemaeker, P.J, Siegal, G, Roe, M, Driscoll, P.C, Abrahams, J.P.A. | | Deposit date: | 1999-02-26 | | Release date: | 1999-10-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the C-terminal SH2 domain of the p85alpha regulatory subunit of phosphoinositide 3-kinase: an SH2 domain mimicking its own substrate.
J.Mol.Biol., 292, 1999
|
|
2KRV
 
 | |
8IZJ
 
 | |
4GCR
 
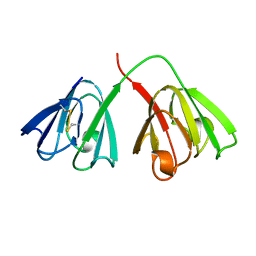 | | STRUCTURE OF THE BOVINE EYE LENS PROTEIN GAMMA-B (GAMMA-II)-CRYSTALLIN AT 1.47 ANGSTROMS | | Descriptor: | GAMMA-B CRYSTALLIN | | Authors: | Slingsby, C, Najmudin, S, Nalini, V, Driessen, H.P.C, Blundell, T.L, Moss, D.S, Lindley, P. | | Deposit date: | 1992-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure of the bovine eye lens protein gammaB(gammaII)-crystallin at 1.47 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
5E0H
 
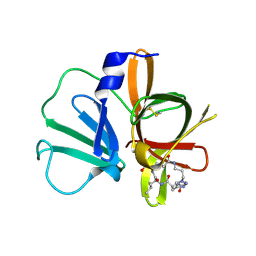 | | 1.95 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic (18-mer) inhibitor | | Descriptor: | (phenylmethyl) ~{N}-[(9~{S},12~{S},15~{S})-9-(hydroxymethyl)-12-(2-methylpropyl)-6,11,14-tris(oxidanylidene)-1,5,10,13,18,19-hexazabicyclo[15.2.1]icosa-17(20),18-dien-15-yl]carbamate, GLYCEROL, Norovirus 3C-like protease | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Weerawarna, P.M, Kim, Y, Kankanamalage, A.C.G, Damalanka, V.C, Lushington, G.H, Alliston, K.R, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2015-09-28 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design and synthesis of triazole-based macrocyclic inhibitors of norovirus protease: Structural, biochemical, spectroscopic, and antiviral studies.
Eur.J.Med.Chem., 119, 2016
|
|
6S00
 
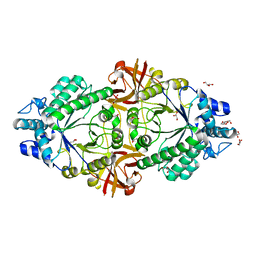 | | Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-acetylneuraminic acid | | Descriptor: | GLYCEROL, N-acetyl-beta-neuraminic acid, TETRAETHYLENE GLYCOL, ... | | Authors: | Bule, P, Blagova, E, Chuzel, L, Taron, C.H, Davies, G.J. | | Deposit date: | 2019-06-13 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inverting family GH156 sialidases define an unusual catalytic motif for glycosidase action.
Nat Commun, 10, 2019
|
|
6RU1
 
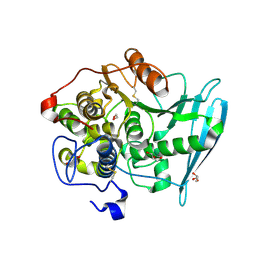 | | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid Um4X | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Ernst, H.A, Mosbech, C, Langkilde, A, Westh, P, Meyer, A, Agger, J.W, Larsen, S. | | Deposit date: | 2019-05-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun, 11, 2020
|
|
6S0E
 
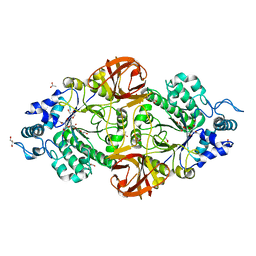 | | Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-Acetyl-2,3-dehydro-2-deoxyneuraminic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, ACETATE ION, ... | | Authors: | Bule, P, Blagova, E, Chuzel, L, Taron, C.H, Davies, G.J. | | Deposit date: | 2019-06-14 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inverting family GH156 sialidases define an unusual catalytic motif for glycosidase action.
Nat Commun, 10, 2019
|
|
1E72
 
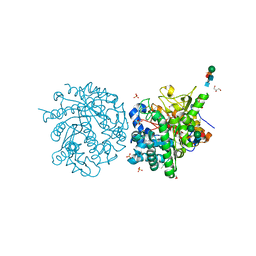 | | Myrosinase from Sinapis alba with bound gluco-hydroximolactam and sulfate or ascorbate | | Descriptor: | (2S,3S,4R,5R)-6-(HYDROXYAMINO)-2-(HYDROXYMETHYL)-2,3,4,5-TETRAHYDROPYRIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Burmeister, W.P. | | Deposit date: | 2000-08-23 | | Release date: | 2001-01-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High resolution X-ray crystallography shows that ascorbate is a cofactor for myrosinase and substitutes for the function of the catalytic base.
J. Biol. Chem., 275, 2000
|
|
5E3O
 
 | |
6NJT
 
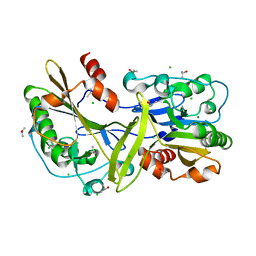 | | Mouse endonuclease G mutant - H97A | | Descriptor: | CHLORIDE ION, Endonuclease G, mitochondrial, ... | | Authors: | Vander Zanden, C.M, Ho, E.N, Czarny, R.S, Robertson, A.B, Ho, P.S. | | Deposit date: | 2019-01-04 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural adaptation of vertebrate endonuclease G for 5-hydroxymethylcytosine recognition and function.
Nucleic Acids Res., 48, 2020
|
|
5E3N
 
 | |
5SWM
 
 | |
6NP3
 
 | | AAC-VIa bound to Gentamicin | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, Aminoglycoside N(3)-acetyltransferase, MAGNESIUM ION | | Authors: | Kumar, P, Cuneo, M.J. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Low-Barrier and Canonical Hydrogen Bonds Modulate Activity and Specificity of a Catalytic Triad.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6HJR
 
 | |
7VP7
 
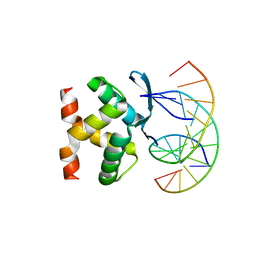 | |
7U67
 
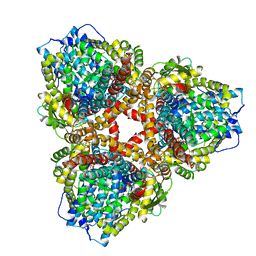 | | Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2 and GTP | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, GUANOSINE-5'-TRIPHOSPHATE, Inhibitor of dGTPase, ... | | Authors: | Klemm, B.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism by which T7 bacteriophage protein Gp1.2 inhibits Escherichia coli dGTPase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6FKE
 
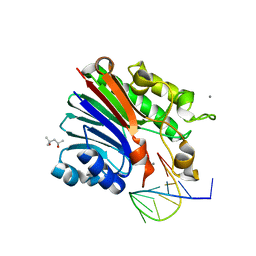 | | Structure of 3' phosphatase NExo (D146N) from Neisseria bound to product DNA hairpin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*TP*AP*GP*CP*GP*AP*AP*GP*CP*TP*A)-3'), Exodeoxyribonuclease III, ... | | Authors: | Silhan, J, Zhao, Q, Boura, E, Thomson, H, Foster, A, Tang, C.M, Freemont, P.S, Baldwin, G.S. | | Deposit date: | 2018-01-23 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structural basis for recognition and repair of the 3'-phosphate by NExo, a base excision DNA repair nuclease from Neisseria meningitidis.
Nucleic Acids Res., 46, 2018
|
|
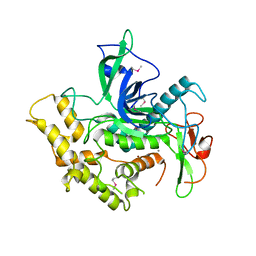
6RIM
 
 | |
