6TK4
 
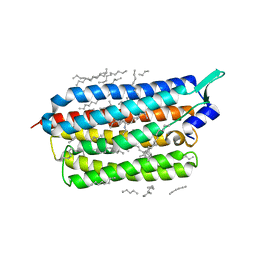 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: 1ns+16ns structure of KR2 with extrapolated, light and dark datasets | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
6TNH
 
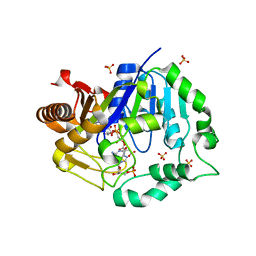 | | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with AMPPcP, dGMP, Asp, Magnesium at 2.21 Angstrom resolution | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ASPARTIC ACID, Adenylosuccinate synthetase, ... | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes.
Science, 372, 2021
|
|
6TK6
 
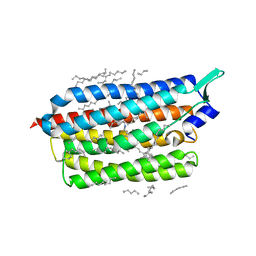 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: Dark structure in neutral conditions with attached light datasets at 800fs, 2ps, 100ps, 1ns, 16ns, 1us, 30us, 150us, 1ms and 20ms | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
6Q8J
 
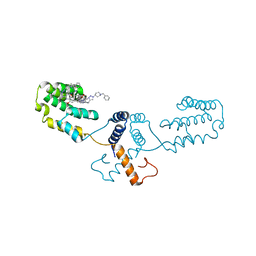 | | Nterminal domain of human SMU1 in complex with LSP641 | | Descriptor: | 1-~{tert}-butyl-3-[6-(3,5-dimethoxyphenyl)-2-[[1-[1-(phenylmethyl)piperidin-4-yl]-1,2,3-triazol-4-yl]methylamino]pyrido[2,3-d]pyrimidin-7-yl]urea, WD40 repeat-containing protein SMU1 | | Authors: | Tengo, L, Le Corre, L, Fournier, G, Ashraf, U, Busca, P, Rameix-Welti, M.-A, Gravier-Pelletier, C, Ruigrok, R.W.H, Jacob, Y, Vidalain, P.-O, Pietrancosta, N, Naffakh, N, McCarthy, A.A, Crepin, T. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Destabilization of the human RED-SMU1 splicing complex as a basis for host-directed antiinfluenza strategy.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5M3D
 
 | | Structural tuning of CD81LEL (space group P31) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen, PHOSPHATE ION | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
6PYK
 
 | |
6NBI
 
 | | Cryo-EM structure of parathyroid hormone receptor type 1 in complex with a long-acting parathyroid hormone analog and G protein | | Descriptor: | CHOLESTEROL, Gs protein alpha subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, L.-H, Ma, S, Sutkeviciute, I, Shen, D.-D, Zhou, X.E, de Waal, P.P, Li, C.-Y, Kang, Y, Clark, L.J, Jean-Alphonse, F.G, White, A.D, Xiao, K, Yang, D, Jiang, Y, Watanabe, T, Gardella, T.J, Melcher, K, Wang, M.-W, Vilardaga, J.-P, Xu, H.E, Zhang, Y. | | Deposit date: | 2018-12-07 | | Release date: | 2019-04-17 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and dynamics of the active human parathyroid hormone receptor-1.
Science, 364, 2019
|
|
7RIX
 
 | | RNA polymerase II elongation complex with hairpin polyamide Py-Im 1, scaffold 2 | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RIW
 
 | | RNA polymerase II elongation complex scaffold 2, without polyamide | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RIM
 
 | | RNA polymerase II elongation complex with hairpin polyamide Py-Im 1, scaffold 1 | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6W1R
 
 | | RT XFEL structure of Photosystem II 150 microseconds after the second illumination at 2.23 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7RIY
 
 | | RNA polymerase II elongation complex with hairpin polyamide Py-Im 1, scaffold 2 soaked with UTP | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RIP
 
 | | RNA polymerase II elongation complex with hairpin polyamide Py-Im 1, scaffold 1 soaked with CTP | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RIQ
 
 | | RNA polymerase II elongation complex scaffold 1 without polyamide | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6XLQ
 
 | | Crystal Structure of the Human BTN3A1 Ectodomain in Complex with the CTX-2026 Fab | | Descriptor: | Butyrophilin subfamily 3 member A1, CTX-2026 Heavy Chain, CTX-2026 Light Chain | | Authors: | Payne, K.K, Mine, J.A, Biswas, S, Chaurio, R.A, Perales-Puchalt, A, Anadon, C.M, Costich, T.L, Harro, C.M, Walrath, J, Ming, Q, Tcyganov, E, Buras, A.L, Rigolizzo, K.E, Mandal, G, Lajoie, J, Ophir, M, Tchou, J, Marchion, D, Luca, V.C, Bobrowicz, P, McLaughlin, B, Eskiocak, U, Schmidt, M, Cubillos-Ruiz, J.R, Rodriguez, P.C, Gabrilovich, D.I, Conejo-Garcia, J.R. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | BTN3A1 governs antitumor responses by coordinating alpha beta and gamma delta T cells.
Science, 369, 2020
|
|
5MMI
 
 | | Structure of the large subunit of the chloroplast ribosome | | Descriptor: | 23S ribosomal RNA, 4.5S ribosomal RNA, 50S ribosomal protein 6, ... | | Authors: | Bieri, P, Leibundgut, M, Saurer, M, Boehringer, D, Ban, N. | | Deposit date: | 2016-12-10 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The complete structure of the chloroplast 70S ribosome in complex with translation factor pY.
EMBO J., 36, 2017
|
|
6XXK
 
 | | Crystal Structure of Human Deoxyhypusine Synthase in complex with spermidine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Wator, E, Wilk, P, Grudnik, P. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Half Way to Hypusine-Structural Basis for Substrate Recognition by Human Deoxyhypusine Synthase.
Biomolecules, 10, 2020
|
|
7RLV
 
 | |
7RLY
 
 | |
6RB2
 
 | | Structure of the (SR)Ca2+-ATPase mutant E340A in the Ca2-E1-CaAMPPCP form | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Clausen, J.D, Montigny, C, Lenoir, G, Arnou, B, Jaxel, C, Moller, J.V, Nissen, P, Andersen, J.P, Le Maire, M, Bublitz, M. | | Deposit date: | 2019-04-09 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.20001125 Å) | | Cite: | The SERCA residue Glu340 mediates interdomain communication that guides Ca 2+ transport.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8GI9
 
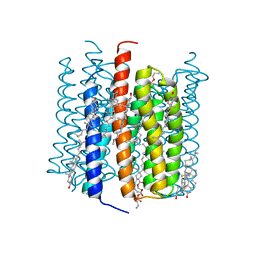 | | Cation channelrhodopsin from Hyphochytrium catenoides (HcCCR) embedded in peptidisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, Cation Channelrhodopsin, ... | | Authors: | Morizumi, T, Kim, K, Li, H, Spudich, J.L, Ernst, O.P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structures of channelrhodopsin paralogs in peptidiscs explain their contrasting K + and Na + selectivities.
Nat Commun, 14, 2023
|
|
8GI8
 
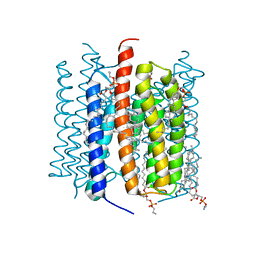 | | Kalium channelrhodopsin 1 from Hyphochytrium catenoides (HcKCR1) embedded in peptidisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, Kalium Channelrhodopsin 1, ... | | Authors: | Morizumi, T, Kim, K, Li, H, Spudich, J.L, Ernst, O.P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structures of channelrhodopsin paralogs in peptidiscs explain their contrasting K + and Na + selectivities.
Nat Commun, 14, 2023
|
|
5MFG
 
 | | Designed armadillo repeat protein YIIIM5AII in complex with peptide (RR)4 | | Descriptor: | (RR)4, CALCIUM ION, YIIIM5AII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
7A94
 
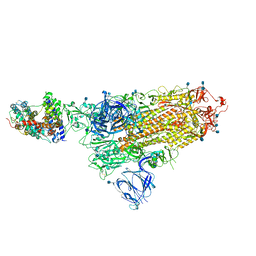 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
6HMS
 
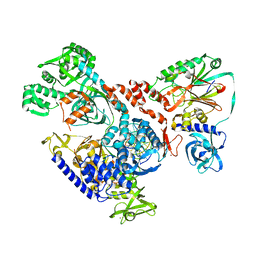 | | Cryo-EM map of DNA polymerase D from Pyrococcus abyssi in complex with DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*TP*C)-3'), DNA (5'-D(P*TP*GP*AP*CP*GP*CP*GP*GP*CP*CP*CP*GP*TP*CP*TP*C)-3'), DNA polymerase II large subunit,DNA polymerase II large subunit, ... | | Authors: | Raia, P, Carroni, M, Sauguet, L. | | Deposit date: | 2018-09-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structure of the DP1-DP2 PolD complex bound with DNA and its implications for the evolutionary history of DNA and RNA polymerases.
PLoS Biol., 17, 2019
|
|
