7PFF
 
 | |
1A3V
 
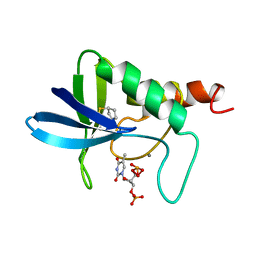 | | STAPHYLOCOCCAL NUCLEASE, CYCLOPENTANE THIOL DISULFIDE TO V23C VARIANT | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Wynn, R, Harkins, P.C, Richards, F.M, Fox, R.O. | | Deposit date: | 1998-01-24 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparison of straight chain and cyclic unnatural amino acids embedded in the core of staphylococcal nuclease.
Protein Sci., 6, 1997
|
|
6N5F
 
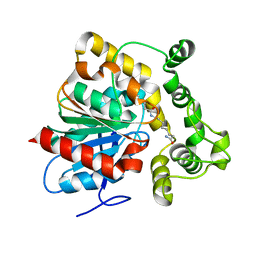 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 3 | | Descriptor: | Epoxide hydrolase TrEH, N-(8-amino-8-oxooctyl)nonanamide | | Authors: | Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
6N5G
 
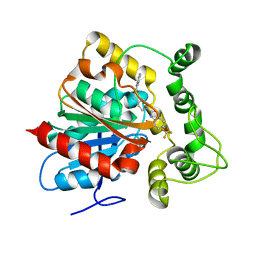 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 2 | | Descriptor: | 4-[(quinolin-3-yl)methyl]-N-[4-(trifluoromethoxy)phenyl]piperidine-1-carboxamide, Epoxide hydrolase TrEH | | Authors: | Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
4V33
 
 | | Crystal structure of the putative polysaccharide deacetylase BA0330 from bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, POLYSACCHARIDE DEACETYLASE-LIKE PROTEIN, ... | | Authors: | Giastas, P, Andreou, A, Bouriotis, V, Eliopoulos, E. | | Deposit date: | 2014-10-16 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Two Putative Polysaccharide Deacetylases are Required for Osmotic Stability and Cell Shape Maintenance in Bacillus Anthracis.
J.Biol.Chem., 290, 2015
|
|
4UUZ
 
 | | MCM2-histone complex | | Descriptor: | DNA REPLICATION LICENSING FACTOR MCM2, HISTONE H3, HISTONE H4 | | Authors: | Richet, N, Liu, D, Legrand, P, Bakail, M, Compper, C, Besle, A, Guerois, R, Ochsenbein, F. | | Deposit date: | 2014-08-01 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insight Into How the Human Helicase Subunit Mcm2 May Act as a Histone Chaperone Together with Asf1 at the Replication Fork.
Nucleic Acids Res., 43, 2015
|
|
4RET
 
 | | Crystal structure of the Na,K-ATPase E2P-digoxin complex with bound magnesium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Gregersen, J.L, Laursen, M, Yatime, L, Nissen, P, Fedosova, N.U. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structures and characterization of digoxin- and bufalin-bound Na+,K+-ATPase compared with the ouabain-bound complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7Q54
 
 | | Single Particle Cryo-EM structure of photosynthetic A4B4-glyceraldehyde 3-phosphate dehydrogenase from Spinacia oleracia. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, Glyceraldehyde-3-phosphate dehydrogenase B, ... | | Authors: | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6MUV
 
 | | The structure of the Plasmodium falciparum 20S proteasome in complex with two PA28 activators | | Descriptor: | 20S proteasome alpha-1 subunit, 20S proteasome alpha-2 subunit, 20S proteasome alpha-3 subunit, ... | | Authors: | Metcalfe, R.D, Xie, S.C, Hanssen, E, Gillett, D.L, Leis, A.P, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2018-10-23 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The structure of the PA28-20S proteasome complex from Plasmodium falciparum and implications for proteostasis.
Nat Microbiol, 4, 2019
|
|
8ELJ
 
 | | SARS-CoV-2 spike glycoprotein in complex with the ICO-hu23 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ICO-hu23 antibody Fab heavy chain, ... | | Authors: | Yee, A.W, Morizumi, T, Kim, K, Kuo, A, Ernst, O.P. | | Deposit date: | 2022-09-24 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Broadly neutralizing humanized SARS-CoV-2 antibody binds to a conserved epitope on Spike and provides antiviral protection through inhalation-based delivery in non-human primates.
Plos Pathog., 19, 2023
|
|
7Q55
 
 | | Single Particle Cryo-EM structure of photosynthetic A8B8 glyceraldehyde-3-phosphate dehydrogenase hexadecamer (major conformer) from Spinacia oleracia. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, Glyceraldehyde-3-phosphate dehydrogenase B, ... | | Authors: | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7Q56
 
 | | Single Particle Cryo-EM structure of photosynthetic A8B8 glyceraldehyde-3-phosphate dehydrogenase (minor conformer) from Spinacia oleracea. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, Glyceraldehyde-3-phosphate dehydrogenase B, ... | | Authors: | Marotta, R, Fermani, S, Sparla, F, Trost, P, Del Giudice, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Unravelling the regulation pathway of photosynthetic AB-GAPDH.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8QJS
 
 | | VHL/Elongin B/Elongin C complex with compound 155 | | Descriptor: | (2S,4R)-1-[(2R)-2-[3-[2-(2-methoxyethoxy)ethoxy]-1,2-oxazol-5-yl]-3-methyl-butanoyl]-N-[(1S)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.191 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
4UUO
 
 | |
8QJR
 
 | | BRG1 bromodomain in complex with VBC via compound 17 | | Descriptor: | (2S,4R)-1-[(2R)-2-[3-[2-[4-[3-[4-[(1R,5S)-3-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]pyridin-2-yl]oxycyclobutyl]oxypiperidin-1-yl]ethoxy]-1,2-oxazol-5-yl]-3-methyl-butanoyl]-N-[(1S)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, CHLORIDE ION, Elongin-B, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
6N1P
 
 | | Dihedral oligomeric complex of GyrA N-terminal fragment with DNA, solved by cryoEM in C2 symmetry | | Descriptor: | DNA (44-MER), DNA gyrase subunit A | | Authors: | Soczek, K.M, Grant, T, Rosenthal, P.B, Mondragon, A. | | Deposit date: | 2018-11-10 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.35 Å) | | Cite: | CryoEM structures of open dimers of Gyrase A in complex with DNA illuminate mechanism of strand passage.
Elife, 7, 2018
|
|
148D
 
 | |
4UTG
 
 | | Burkholderia pseudomallei heptokinase WcbL,AMPPNP (ATP analogue) complex. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Vivoli, M, Isupov, M.N, Nicholas, R, Hill, A, Scott, A, Kosma, P, Prior, J, Harmer, N.J. | | Deposit date: | 2014-07-21 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Unraveling the B.Pseudomallei Heptokinase Wcbl: From Structure to Drug Discovery.
Chem.Biol., 22, 2015
|
|
1A3U
 
 | | STAPHYLOCOCCAL NUCLEASE, CYCLOHEXANE THIOL DISULFIDE TO V23C VARIANT | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Wynn, R, Harkins, P.C, Richards, F.M, Fox, R.O. | | Deposit date: | 1998-01-24 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Comparison of straight chain and cyclic unnatural amino acids embedded in the core of staphylococcal nuclease.
Protein Sci., 6, 1997
|
|
6N9Y
 
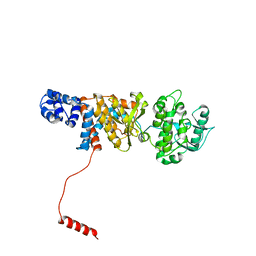 | | Atomic structure of Non-Structural protein 1 of bluetongue virus | | Descriptor: | Non-structural protein 1 | | Authors: | Kerviel, A, Ge, P, Lai, M, Jih, J, Boyce, M, Zhang, X, Zhou, Z.H, Roy, P. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Atomic structure of the translation regulatory protein NS1 of bluetongue virus.
Nat Microbiol, 4, 2019
|
|
3CKZ
 
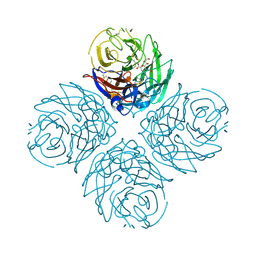 | | N1 Neuraminidase H274Y + Zanamivir | | Descriptor: | CALCIUM ION, Neuraminidase, ZANAMIVIR | | Authors: | Colllins, P, Haire, L.F, Lin, Y.P, Liu, J, Russell, R.J, Walker, P.A, Skehel, J.J, Martin, S.R, Hay, A.J, Gamblin, S.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-05-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants.
Nature, 453, 2008
|
|
8PIL
 
 | | E. coli transcription complex paused at ops site and bound to RfaH and NusA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuber, P.K, Said, N, Hilal, T, Loll, B, Wahl, M.C, Knauer, S.H. | | Deposit date: | 2023-06-22 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Concerted transformation of a hyper-paused transcription complex and its reinforcing protein.
Nat Commun, 15, 2024
|
|
8PIM
 
 | | fully recruited RfaH bound to E. coli transcription complex paused at ops site (not complementary scaffold) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuber, P.K, Said, N, Hilal, T, Loll, B, Wahl, M.C, Knauer, S.H. | | Deposit date: | 2023-06-22 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Concerted transformation of a hyper-paused transcription complex and its reinforcing protein.
Nat Commun, 15, 2024
|
|
6YL3
 
 | | High resolution cryo-EM structure of urease from the pathogen Yersinia enterocolitica | | Descriptor: | NICKEL (II) ION, Urease subunit alpha, Urease subunit beta, ... | | Authors: | Righetto, R.D, Anton, L, Adaixo, R, Jakob, R, Zivanov, J, Mahi, M.A, Ringler, P, Schwede, T, Maier, T, Stahlberg, H. | | Deposit date: | 2020-04-06 | | Release date: | 2020-05-06 | | Last modified: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | High-resolution cryo-EM structure of urease from the pathogen Yersinia enterocolitica.
Nat Commun, 11, 2020
|
|
1AGS
 
 | |
