3GAR
 
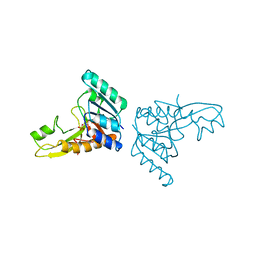 | | A PH-DEPENDENT STABLIZATION OF AN ACTIVE SITE LOOP OBSERVED FROM LOW AND HIGH PH CRYSTAL STRUCTURES OF MUTANT MONOMERIC GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, PHOSPHATE ION | | Authors: | Su, Y, Yamashita, M.M, Greasley, S.E, Mullen, C.A, Shim, J.H, Jennings, P.A, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A pH-dependent stabilization of an active site loop observed from low and high pH crystal structures of mutant monomeric glycinamide ribonucleotide transformylase at 1.8 to 1.9 A.
J.Mol.Biol., 281, 1998
|
|
3P43
 
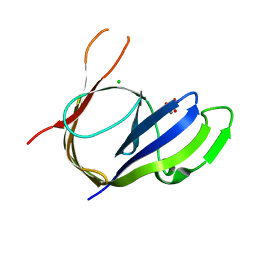 | | Structure and Activities of Archaeal Members of the LigD 3' Phosphoesterase DNA Repair Enzyme Superfamily | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Smith, P, Nair, P.A, Das, U, Shuman, S. | | Deposit date: | 2010-10-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures and activities of archaeal members of the LigD 3'-phosphoesterase DNA repair enzyme superfamily.
Nucleic Acids Res., 39, 2011
|
|
4JA4
 
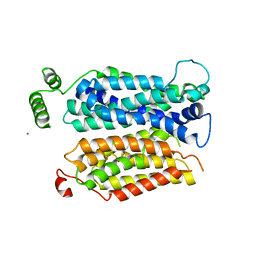 | | Inward open conformation of the xylose transporter XylE from E. coli | | Descriptor: | CADMIUM ION, D-xylose-proton symporter | | Authors: | Quistgaard, E.M, Low, C, Moberg, P, Tresaugues, L, Nordlund, P. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural basis for substrate transport in the GLUT-homology family of monosaccharide transporters.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1QPI
 
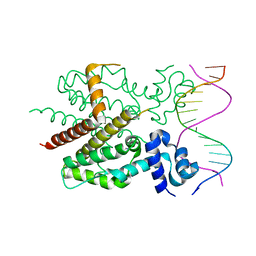 | | CRYSTAL STRUCTURE OF TETRACYCLINE REPRESSOR/OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*CP*CP*TP*AP*TP*CP*AP*AP*TP*GP*AP*TP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*AP*TP*CP*AP*TP*TP*GP*AP*TP*AP*GP*G)-3'), IMIDAZOLE, ... | | Authors: | Orth, P, Schnappinger, D, Hillen, W, Saenger, W, Hinrichs, W. | | Deposit date: | 1999-05-25 | | Release date: | 2000-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system.
Nat.Struct.Biol., 7, 2000
|
|
4JKT
 
 | | Crystal structure of mouse Glutaminase C, BPTES-bound form | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide) | | Authors: | Fornezari, C, Ferreira, A.P.S, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2013-03-11 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Active Glutaminase C Self-assembles into a Supratetrameric Oligomer That Can Be Disrupted by an Allosteric Inhibitor.
J.Biol.Chem., 288, 2013
|
|
4NWR
 
 | | Computationally Designed Two-Component Self-Assembling Tetrahedral Cage T33-28 | | Descriptor: | Macrophage migration inhibitory factor-like protein, integron gene cassette protein | | Authors: | McNamara, D.E, King, N.P, Bale, J.B, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2013-12-06 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Accurate design of co-assembling multi-component protein nanomaterials.
Nature, 510, 2014
|
|
354D
 
 | | Structure of loop E FROM E. coli 5S RRNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*CP*CP*GP*AP*UP*GP*GP*UP*AP*GP*UP*G)-3'), RNA-DNA (5'-R(*GP*CP*GP*AP*GP*AP*GP*UP*AP*)-D(*DGP(S)*)-R(*GP*C)-3') | | Authors: | Correll, C.C, Freeborn, B, Moore, P.B, Steitz, T.A. | | Deposit date: | 1997-10-01 | | Release date: | 1997-11-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain.
Cell(Cambridge,Mass.), 91, 1997
|
|
2Q5B
 
 | | High resolution structure of Plastocyanin from Phormidium laminosum | | Descriptor: | ACETATE ION, COPPER (II) ION, POTASSIUM ION, ... | | Authors: | Fromme, R, Bukhman-DeRuyter, Y.S, Grotjohann, I, Mi, H, Fromme, P. | | Deposit date: | 2007-05-31 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Plastocyanin at high resolution from Phormidium laminosum and the double mutant D44A, D45A
To be Published
|
|
2Q8J
 
 | | Crystal Structure of the complex of C-lobe of bovine lactoferrin with Mannitol and Mannose at 2.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Mir, R, Jain, R, Sinha, M, Singh, N, Sharma, S, Kaur, P, Bhushan, A, Singh, T.P. | | Deposit date: | 2007-06-11 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of the complex of C-lobe of bovine lactoferrin with Mannitol and Mannose at 2.7 A resolution
To be Published
|
|
4JQL
 
 | | Synthesis of Benzoquinone-Ansamycin-Inspired Macrocyclic Lactams from Shikimic Acid | | Descriptor: | Heat shock protein HSP 90-alpha, MAGNESIUM ION, valerjesomycin | | Authors: | Jeso, V, Iqbal, S, Hernandez, P, Cameron, M.D, Park, H, Lograsso, P.V, Micalizio, G.C. | | Deposit date: | 2013-03-20 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Synthesis of benzoquinone ansamycin-inspired macrocyclic lactams from shikimic Acid.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
1J6Z
 
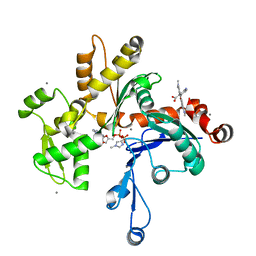 | | UNCOMPLEXED ACTIN | | Descriptor: | ACTIN ALPHA 1, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Otterbein, L.R, Graceffa, P, Dominguez, R. | | Deposit date: | 2001-05-15 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The crystal structure of uncomplexed actin in the ADP state.
Science, 293, 2001
|
|
1F84
 
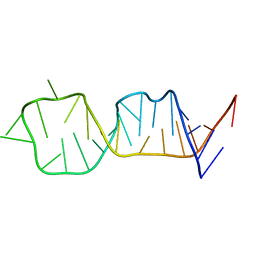 | | SOLUTION STRUCTURE OF HCV IRES RNA DOMAIN IIID | | Descriptor: | HCV-1B IRES RNA DOMAIN IIID | | Authors: | Lukavsky, P.J, Otto, G.A, Lancaster, A.M, Sarnow, P, Puglisi, J.D. | | Deposit date: | 2000-06-28 | | Release date: | 2000-11-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function.
Nat.Struct.Biol., 7, 2000
|
|
1QBQ
 
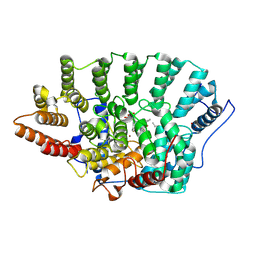 | | STRUCTURE OF RAT FARNESYL PROTEIN TRANSFERASE COMPLEXED WITH A CVIM PEPTIDE AND ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID. | | Descriptor: | ACETATE ION, ACETYL-CYS-VAL-ILE-SELENOMET-COOH PEPTIDE, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, ... | | Authors: | Strickland, C.L, Windsor, W.T, Syto, R, Wang, L, Bond, R, Wu, Z, Schwartz, J, Le, H.V, Beese, L.S, Weber, P.C. | | Deposit date: | 1999-04-27 | | Release date: | 1999-06-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of farnesyl protein transferase complexed with a CaaX peptide and farnesyl diphosphate analogue
Biochemistry, 37, 1998
|
|
1FCY
 
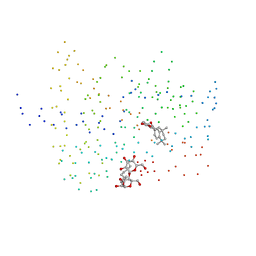 | | ISOTYPE SELECTIVITY OF THE HUMAN RETINOIC ACID NUCLEAR RECEPTOR HRAR: THE COMPLEX WITH THE RARBETA/GAMMA-SELECTIVE RETINOID CD564 | | Descriptor: | 6-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTALENE-2-CARBONYL)-NAPHTALENE-2-CARBOXYLIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-1 | | Authors: | Klaholz, B.P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-07-19 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for isotype selectivity of the human retinoic acid nuclear receptor.
J.Mol.Biol., 302, 2000
|
|
3Q38
 
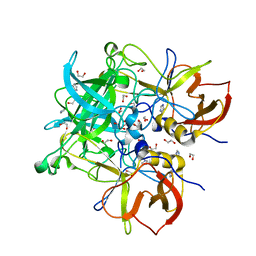 | | Crystal structure of P domain from norwalk virus strain vietnam 026 in complex with HBGA type B (triglycan) | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, IMIDAZOLE, ... | | Authors: | Hansman, G.S, Biertumpfel, C, Chen, L, Georgiev, I, McLellan, J.S, Katayama, K, Kwong, P.D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability.
J.Virol., 85, 2011
|
|
1TGG
 
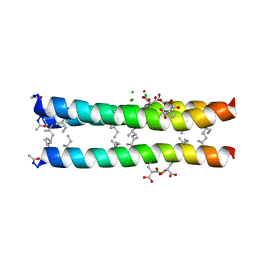 | | RH3 DESIGNED RIGHT-HANDED COILED COIL TRIMER | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, right-handed coiled coil trimer | | Authors: | Plecs, J.J, Harbury, P.B, Kim, P.S, Alber, T. | | Deposit date: | 2004-05-28 | | Release date: | 2004-10-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural test of the parameterized-backbone method for protein design
J.Mol.Biol., 342, 2004
|
|
1GVX
 
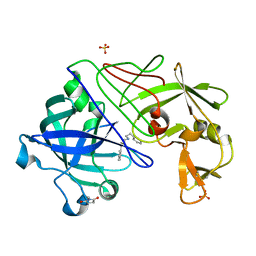 | | Endothiapepsin complexed with H256 | | Descriptor: | ENDOTHIAPEPSIN, INHIBITOR H256, SULFATE ION | | Authors: | Coates, L, Erskine, P.T, Crump, M.P, Wood, S.P, Cooper, J.B. | | Deposit date: | 2002-02-27 | | Release date: | 2002-07-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Five Atomic Resolution Structures of Endothiapepsin Inhibitor Complexes: Implications for the Aspartic Proteinase Mechanism
J.Mol.Biol., 318, 2002
|
|
1LPP
 
 | |
1X9N
 
 | | Crystal Structure of Human DNA Ligase I bound to 5'-adenylated, nicked DNA | | Descriptor: | 5'-phosphorylated DNA, ADENOSINE MONOPHOSPHATE, DNA ligase I, ... | | Authors: | Pascal, J.M, O'Brien, P.J, Tomkinson, A.E, Ellenberger, T. | | Deposit date: | 2004-08-23 | | Release date: | 2004-11-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human DNA ligase I completely encircles and partially unwinds nicked DNA.
Nature, 432, 2004
|
|
4FAL
 
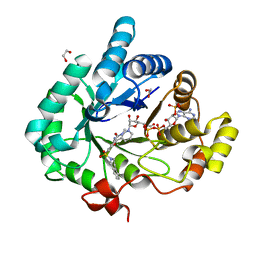 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 in complex with 3-((3,4-dihydroisoquinolin-2(1H)-yl)sulfonyl)-N-methylbenzamide (80) | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,4-dihydroisoquinolin-2(1H)-ylsulfonyl)-N-methylbenzamide, Aldo-keto reductase family 1 member C3, ... | | Authors: | Turnbull, A.P, Jamieson, S.M.F, Brooke, D.G, Heinrich, D, Atwell, G.J, Silva, S, Hamilton, E.J, Rigoreau, L.J.M, Trivier, E, Soudy, C, Samlal, S.S, Owen, P.J, Schroeder, E, Raynham, T, Flanagan, J.U, Denny, W.A. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-(3,4-Dihydroisoquinolin-2(1H)-ylsulfonyl)benzoic acids; a New Class of Highly Potent and Selective Inhibitors of the Type 5 17-beta-hydroxysteroid Dehydrogenase AKR1C3
J.Med.Chem., 55, 2012
|
|
4NYA
 
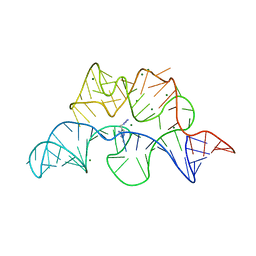 | | Crystal structure of the E. coli thiM riboswitch in complex with 5-(azidomethyl)-2-methylpyrimidin-4-amine | | Descriptor: | 5-(azidomethyl)-2-methylpyrimidin-4-amine, MAGNESIUM ION, thiM TPP riboswitch | | Authors: | Warner, K.D, Homan, P, Weeks, K.M, Smith, A.G, Abell, C, Ferre-D'Amare, A.R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Validating Fragment-Based Drug Discovery for Biological RNAs: Lead Fragments Bind and Remodel the TPP Riboswitch Specifically.
Chem.Biol., 21, 2014
|
|
1LDZ
 
 | |
1XD3
 
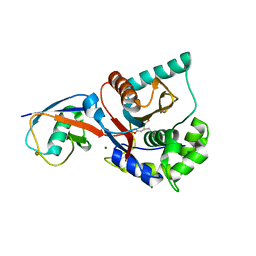 | | Crystal structure of UCHL3-UbVME complex | | Descriptor: | MAGNESIUM ION, METHYL 4-AMINOBUTANOATE, UBC protein, ... | | Authors: | Misaghi, S, Galardy, P.J, Meester, W.J.N, Ovaa, H, Ploegh, H.L, Gaudet, R. | | Deposit date: | 2004-09-03 | | Release date: | 2004-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the Ubiquitin Hydrolase UCH-L3 Complexed with a Suicide Substrate
J.Biol.Chem., 280, 2005
|
|
1LPO
 
 | |
4F8H
 
 | | X-ray Structure of the Anesthetic Ketamine Bound to the GLIC Pentameric Ligand-gated Ion Channel | | Descriptor: | (R)-ketamine, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Proton-gated ion channel, ... | | Authors: | Pan, J.J, Chen, Q, Willenbring, D, Kong, X.P, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2012-05-17 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structure of the Pentameric Ligand-Gated Ion Channel GLIC Bound with Anesthetic Ketamine.
Structure, 20, 2012
|
|
