7SC1
 
 | |
6VOL
 
 | | Chloroplast ATP synthase (R2, CF1FO) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
5CDN
 
 | | 2.8A structure of etoposide with S.aureus DNA gyrase and DNA | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP**GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*C*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*C)-3'), ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
6VM4
 
 | | Chloroplast ATP synthase (C2, CF1FO) | | Descriptor: | ATP synthase delta chain, chloroplastic, ATP synthase epsilon chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.08 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6VON
 
 | | Chloroplast ATP synthase (R1, CF1FO) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
5JEV
 
 | | del-[Ru(phen)2(dppz]2+ bound to d(TCGGCGCCGA) with Cobalt hexammine | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*GP*A)-3'), Delta-Ru(phen)2(dppz) complex | | Authors: | Hall, J.P, Cardin, C.J. | | Deposit date: | 2016-04-19 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Delta chirality ruthenium 'light-switch' complexes can bind in the minor groove of DNA with five different binding modes.
Nucleic Acids Res., 44, 2016
|
|
6R7Y
 
 | | CryoEM structure of calcium-bound human TMEM16K / Anoctamin 10 in detergent (low Ca2+, closed form) | | Descriptor: | Anoctamin-10, CALCIUM ION | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
6RBD
 
 | | State 1 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Mitterer, V, Ferreira-Cerca, S, Murat, G, Enne, T, Rinaldi, D, Weigl, S, Omanic, H, Gleizes, P.E, Kressler, D, Pertschy, B, Plisson-Chastang, C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism.
Nat Commun, 10, 2019
|
|
7NW1
 
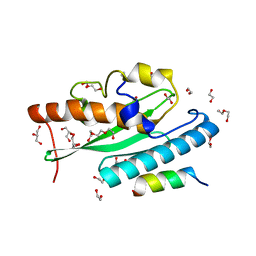 | | Crystal structure of UFC1 in complex with UBA5 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-16 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
5MWF
 
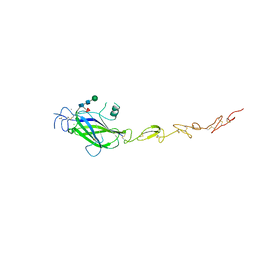 | | Human Jagged2 C2-EGF2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
6RAX
 
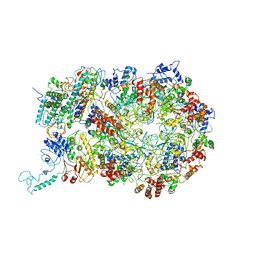 | | D. melanogaster CMG-DNA, State 1B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, AT18545p, ... | | Authors: | Eickhoff, P, Martino, F, Costa, A. | | Deposit date: | 2019-04-08 | | Release date: | 2019-09-11 | | Last modified: | 2019-09-18 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Molecular Basis for ATP-Hydrolysis-Driven DNA Translocation by the CMG Helicase of the Eukaryotic Replisome.
Cell Rep, 28, 2019
|
|
6RBE
 
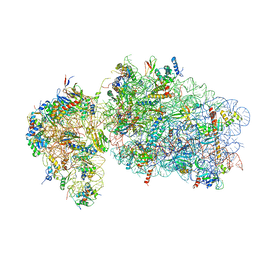 | | State 2 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Mitterer, V, Ferreira-Cerca, S, Murat, G, Enne, T, Rinaldi, D, Weigl, S, Omanic, H, Gleizes, P.E, Kressler, D, Pertschy, B, Plisson-Chastang, C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism.
Nat Commun, 10, 2019
|
|
4XWG
 
 | | Crystal Structure of LCAT (C31Y) in complex with Fab1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1 Heavy Chain, Fab1 Light Chain, ... | | Authors: | Piper, D.E, Walker, N.P.C, Romanow, W.G, Thibault, S.T. | | Deposit date: | 2015-01-28 | | Release date: | 2015-07-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The high-resolution crystal structure of human LCAT.
J.Lipid Res., 56, 2015
|
|
5JAR
 
 | |
7SYN
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 8(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, HCV IRES, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
6Y7V
 
 | | Crystal structure of the KDEL receptor bound to HDEL peptide at pH 6.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON DIOXIDE, ER lumen protein-retaining receptor 2, ... | | Authors: | Braeuer, P, Newstead, S. | | Deposit date: | 2020-03-02 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | A signal capture and proofreading mechanism for the KDEL-receptor explains selectivity and dynamic range in ER retrieval.
Elife, 10, 2021
|
|
8P60
 
 | | Spraguea lophii ribosome dimer | | Descriptor: | 40S Ribosomal protein S19, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Gil Diez, P, McLaren, M, Isupov, M.N, Daum, B, Conners, R, Williams, B. | | Deposit date: | 2023-05-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (14.3 Å) | | Cite: | CryoEM reveals that ribosomes in microsporidian spores are locked in a dimeric hibernating state.
Nat Microbiol, 8, 2023
|
|
7SYW
 
 | | Structure of the wt IRES eIF5B-containing 48S initiation complex, closed conformation. Structure 15(wt) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYG
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 1(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYM
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 7(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Comprehensive structural overview of the HCV IRES-mediated translation initiation pathway
To Be Published
|
|
6Y08
 
 | | Mouse thymidylate synthase cocrystallized with dUMP and soaked in sulfamethoxazole | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Sulfamethoxazole, Thymidylate synthase | | Authors: | Maj, P, Jarmula, A, Wilk, P, Weiss, M.S, Rode, W. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Mouse thymidylate synthase cocrystallized with dUMP and soaked in sulfamethoxazole
To Be Published
|
|
6VOH
 
 | | Chloroplast ATP synthase (O1, CF1FO) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
7MCC
 
 | | Crystal structure of an AI-designed TIM-barrel F2C | | Descriptor: | AI-designed TIM-barrel F2C, SULFATE ION | | Authors: | Mathews, I.I, Anand-Achim, N, Perez, C.P, Huang, P.S. | | Deposit date: | 2021-04-02 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Protein sequence design with a learned potential.
Nat Commun, 13, 2022
|
|
6VWS
 
 | | Hexamer of Helical HIV capsid by RASTR method | | Descriptor: | HIV capsid protein | | Authors: | Zhao, H, Iqbal, N, Asturias, F, Kvaratskhelia, M, Vanblerkom, P. | | Deposit date: | 2020-02-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.08 Å) | | Cite: | Structural and mechanistic bases for a potent HIV-1 capsid inhibitor.
Science, 370, 2020
|
|
4YGM
 
 | | Vaccinia virus his-D4/A20(1-50) in complex with uracil | | Descriptor: | DNA polymerase processivity factor component A20, SULFATE ION, URACIL, ... | | Authors: | Tarbouriech, N, Iseni, F, Burmeister, W.P. | | Deposit date: | 2015-02-26 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Vaccinia Virus Uracil-DNA Glycosylase in Complex with DNA.
J.Biol.Chem., 290, 2015
|
|
