1CKW
 
 | | CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR: SOLUTION STRUCTURES OF PEPTIDES BASED ON THE PHE508 REGION, THE MOST COMMON SITE OF DISEASE-CAUSING DELTA-F508 MUTATION | | Descriptor: | PROTEIN (CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR (CFTR)) | | Authors: | Massiah, M.A, Ko, Y.H, Pedersen, P.L, Mildvan, A.S. | | Deposit date: | 1999-04-26 | | Release date: | 1999-05-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Cystic fibrosis transmembrane conductance regulator: solution structures of peptides based on the Phe508 region, the most common site of disease-causing DeltaF508 mutation.
Biochemistry, 38, 1999
|
|
2E9C
 
 | | E. coli undecaprenyl pyrophosphate synthase in complex with BPH-675 | | Descriptor: | 1-HYDROXY-2-[3'-(NAPHTHALENE-2-SULFONYLAMINO)-BIPHENYL-3-YL]ETHYLIDENE-1,1-BISPHOSPHONIC ACID, Undecaprenyl pyrophosphate synthetase | | Authors: | Guo, R.T, Ko, T.P, Cao, R, Liang, P.H, Oldfield, E, Wang, A.H.J. | | Deposit date: | 2007-01-24 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3NL3
 
 | | The Crystal Structure of Candida glabrata THI6, a Bifunctional Enzyme involved in Thiamin Biosyhthesis of Eukaryotes | | Descriptor: | MAGNESIUM ION, THIAMIN PHOSPHATE, Thiamine biosynthetic bifunctional enzyme | | Authors: | Paul, D, Chatterjee, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.007 Å) | | Cite: | Domain Organization in Candida glabrata THI6, a Bifunctional Enzyme Required for Thiamin Biosynthesis in Eukaryotes .
Biochemistry, 49, 2010
|
|
4AGU
 
 | | CRYSTAL STRUCTURE OF THE HUMAN CDKL1 KINASE DOMAIN | | Descriptor: | CYCLIN-DEPENDENT KINASE-LIKE 1, N-(5-{[(2S)-4-amino-2-(3-chlorophenyl)butanoyl]amino}-1H-indazol-3-yl)benzamide | | Authors: | Canning, P, Sharpe, T.D, Allerston, C, Savitsky, P, Pike, A.C.W, Muniz, J.R.C, Chaikuad, A, Kuo, K, Burgess-Brown, N, Ayinampudi, V, Zhang, Y, Thangaratnarajah, C, Ugochukwu, E, Vollmar, M, Krojer, T, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Knapp, S, Bullock, A. | | Deposit date: | 2012-01-31 | | Release date: | 2012-02-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CDKL Family Kinases Have Evolved Distinct Structural Features and Ciliary Function.
Cell Rep, 22, 2018
|
|
3DHR
 
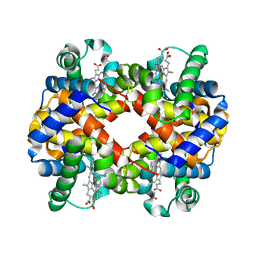 | | Crystal Structure Determination of Methemoglobin from Pigeon at 2 Angstrom Resolution (Columba livia) | | Descriptor: | Hemoglobin subunit alpha-A, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sathya Moorthy, P, Neelagandan, K, Balasubramanian, M, Ponnuswamy, M.N. | | Deposit date: | 2008-06-18 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Purification, Crystallization and X-ray Diffraction Studies of Pigeon Hemoglobin
To be Published
|
|
5MOT
 
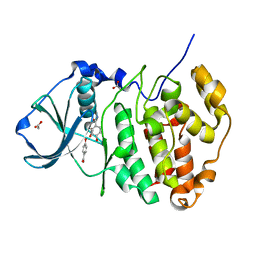 | | Crystal structure of CK2alpha with ZT0627 bound | | Descriptor: | 4-HYDROXYBENZAMIDE, ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Georgiou, K, Iegre, J, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2016-12-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
5MRV
 
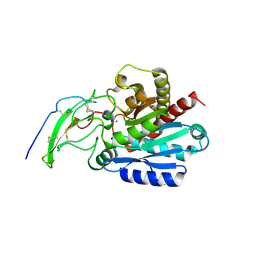 | | Crystal structure of human carboxypeptidase O in complex with NvCI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carboxypeptidase O, Metallocarboxypeptidase inhibitor, ... | | Authors: | Garcia-Pardo, J, Garcia-Guerrero, M.C, Fernandez-Alvarez, R, Lyons, P, Aviles, F.X, Lorenzo, J, Reverter, D. | | Deposit date: | 2016-12-27 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Crystal structure and mechanism of human carboxypeptidase O: Insights into its specific activity for acidic residues.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2PXD
 
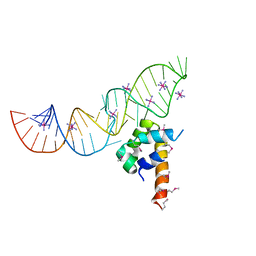 | | Variant 1 of Ribonucleoprotein Core of the E. Coli Signal Recognition Particle | | Descriptor: | 4.5 S RNA, COBALT HEXAMMINE(III), Signal recognition particle protein | | Authors: | Keel, A.Y, Rambo, R.P, Batey, R.T, Kieft, J.S. | | Deposit date: | 2007-05-14 | | Release date: | 2007-08-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A General Strategy to Solve the Phase Problem in RNA Crystallography.
Structure, 15, 2007
|
|
2YNO
 
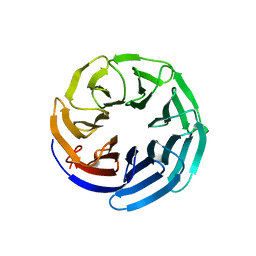 | | yeast betaprime COP 1-304H6 | | Descriptor: | COATOMER SUBUNIT BETA', POLY ALA | | Authors: | Jackson, L.P, Lewis, M, Kent, H.M, Edeling, M.A, Evans, P.R, Duden, R, Owen, D.J. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Recognition of Dilysine Trafficking Motifs by Copi.
Dev.Cell, 23, 2012
|
|
2PXP
 
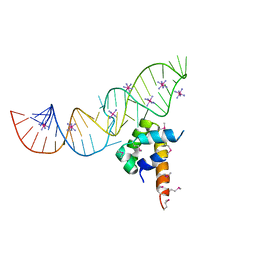 | | Variant 13 of Ribonucleoprotein Core of the E. Coli Signal Recognition Particle | | Descriptor: | 4.5 S RNA, COBALT HEXAMMINE(III), Signal recognition particle protein | | Authors: | Keel, A.Y, Rambo, R.P, Batey, R.T, Kieft, J.S. | | Deposit date: | 2007-05-14 | | Release date: | 2007-08-07 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A General Strategy to Solve the Phase Problem in RNA Crystallography.
Structure, 15, 2007
|
|
2JLS
 
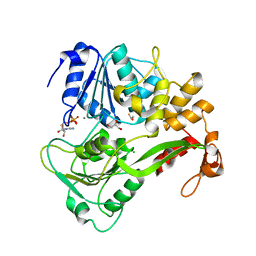 | | Dengue virus 4 NS3 helicase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
1PSI
 
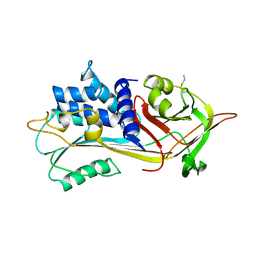 | |
2WSY
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE | | Authors: | Schneider, T.R, Gerhardt, E, Lee, M, Liang, P.-H, Anderson, K.S, Schlichting, I. | | Deposit date: | 1998-02-18 | | Release date: | 1999-03-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Loop closure and intersubunit communication in tryptophan synthase.
Biochemistry, 37, 1998
|
|
1CVQ
 
 | | SOLUTION STRUCTURE OF THE ANALOGUE RETRO-INVERSO MGREGRIGGC IN CONTACT WITH THE MONOCLONAL ANTIBODY MAB 4X11, NMR, 7 STRUCTURES | | Descriptor: | HISTONE H3 | | Authors: | Phan Chan Du, A, Petit, M.C, Guichard, G, Briand, J.P, Muller, S, Cung, M.T. | | Deposit date: | 1999-08-24 | | Release date: | 1999-09-02 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of antibody-bound peptides and retro-inverso analogues. A transferred nuclear Overhauser effect spectroscopy and molecular dynamics approach.
Biochemistry, 40, 2001
|
|
2Y75
 
 | | The Structure of CymR (YrzC) the Global Cysteine Regulator of B. subtilis | | Descriptor: | CHLORIDE ION, HTH-TYPE TRANSCRIPTIONAL REGULATOR CYMR, SULFATE ION | | Authors: | Shepard, W, Soutourina, O, Courtois, E, England, P, Haouz, A, Martin-Verstraete, I. | | Deposit date: | 2011-01-28 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights Into the Rrf2 Repressor Family - the Structure of Cymr, the Global Cysteine Regulator of Bacillus Subtilis.
FEBS J., 278, 2011
|
|
1KRA
 
 | |
3PQ7
 
 | | Structure of I274C variant of E. coli KatE[] - Images 31-36 | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
5MTL
 
 | | Crystal structure of an amyloidogenic light chain | | Descriptor: | light chain dimer,IGL@ protein,IGL@ protein | | Authors: | Oberti, L, Rognoni, P, Russo, R, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-10 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
2WHB
 
 | | Truncation and Optimisation of Peptide Inhibitors of CDK2, Cyclin A Through Structure Guided Design | | Descriptor: | ARG-ARG-L3O-PFF, CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2 | | Authors: | Kontopidis, G, Andrews, M.J, McInnes, C, Plater, A, Innes, L, Renachowski, S, Cowan, A, Fischer, P.M. | | Deposit date: | 2009-05-03 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Truncation and optimisation of peptide inhibitors of cyclin-dependent kinase 2-cyclin a through structure-guided design.
Chemmedchem, 4, 2009
|
|
5WAN
 
 | | Crystal Structure of a flavoenzyme RutA in the pyrimidine catabolic pathway | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Pyrimidine monooxygenase RutA, ... | | Authors: | Zhang, Y, Mukherjee, T, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2017-06-26 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Catalysis of a flavoenzyme-mediated amide hydrolysis.
J. Am. Chem. Soc., 132, 2010
|
|
1CKZ
 
 | | CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR: SOLUTION STRUCTURES OF PEPTIDES BASED ON THE PHE508 REGION, THE MOST COMMON SITE OF DISEASE-CAUSING DELTA-F508 MUTATION | | Descriptor: | PROTEIN (CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR (CFTR)) | | Authors: | Massiah, M.A, Ko, Y.H, Pedersen, P.L, Mildvan, A.S. | | Deposit date: | 1999-04-26 | | Release date: | 1999-05-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Cystic fibrosis transmembrane conductance regulator: solution structures of peptides based on the Phe508 region, the most common site of disease-causing DeltaF508 mutation.
Biochemistry, 38, 1999
|
|
1KZN
 
 | | Crystal Structure of E. coli 24kDa Domain in Complex with Clorobiocin | | Descriptor: | CLOROBIOCIN, DNA GYRASE SUBUNIT B | | Authors: | Lafitte, D, Lamour, V, Tsvetkov, P.O, Makarov, A.A, Klich, M, Deprez, P, Moras, D, Briand, C, Gilli, R. | | Deposit date: | 2002-02-07 | | Release date: | 2002-06-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA gyrase interaction with coumarin-based inhibitors: the role of the hydroxybenzoate isopentenyl moiety and the 5'-methyl group of the noviose.
Biochemistry, 41, 2002
|
|
4D81
 
 | | Metallosphera sedula Vps4 crystal structure | | Descriptor: | AAA ATPase, central domain protein, ADENOSINE-5'-DIPHOSPHATE | | Authors: | Caillat, C, Macheboeuf, P, Wu, Y, McCarthy, A.A, Boeri-Erba, E, Effantin, G, Gottlinger, H.G, Weissenhorn, W, Renesto, P. | | Deposit date: | 2014-12-02 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Asymmetric Ring Structure of Vps4 Required for Escrt-III Disassembly.
Nat.Commun., 6, 2015
|
|
5MWJ
 
 | | Structure Enabled Discovery of a Stapled Peptide Inhibitor to Target the Oncogenic Transcriptional Repressor TLE1 | | Descriptor: | DIMETHYL SULFOXIDE, Transducin-like enhancer protein 1, pepide inhibtor | | Authors: | McGrath, S, Tortorici, M, Vidler, L, Drouin, L, Westwood, I, Gimeson, P, Van Montfort, R, Hoelder, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure-Enabled Discovery of a Stapled Peptide Inhibitor to Target the Oncogenic Transcriptional Repressor TLE1.
Chemistry, 23, 2017
|
|
1YDE
 
 | | Crystal Structure of Human Retinal Short-Chain Dehydrogenase/Reductase 3 | | Descriptor: | Retinal dehydrogenase/reductase 3 | | Authors: | Lukacik, P, Bunkozci, G, Kavanagh, K, Sundstrom, M, Arrowsmith, C, Edwards, A, von Delft, F, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2004-12-23 | | Release date: | 2005-01-18 | | Last modified: | 2012-03-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biochemical characterization of human orphan DHRS10 reveals a novel cytosolic enzyme with steroid dehydrogenase activity.
Biochem.J., 402, 2007
|
|
