5LK8
 
 | |
2MXF
 
 | |
2Y02
 
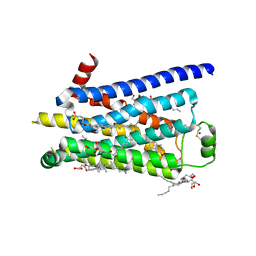 | | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND AGONIST CARMOTEROL | | Descriptor: | BETA-1 ADRENERGIC RECEPTOR, CARMOTEROL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Warne, A, Moukhametzianov, R, Baker, J.G, Nehme, R, Edwards, P.C, Leslie, A.G.W, Schertler, G.F.X, Tate, C.G. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structural Basis for Agonist and Partial Agonist Action on a Beta1-Adrenergic Receptor
Nature, 469, 2011
|
|
5TIO
 
 | | Crystal Structure of Human Glycine Receptor alpha-3 Bound to AM-3607 | | Descriptor: | (3S,3aS,9bS)-2-[(2H-1,3-benzodioxol-5-yl)sulfonyl]-3,5-dimethyl-1,2,3,3a,5,9b-hexahydro-4H-pyrrolo[3,4-c][1,6]naphthyridin-4-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Shaffer, P.L, Huang, X, Chen, H. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal Structures of Human GlyRa3 Bound to a Novel Class of Potentiators with Efficacy in a Mouse Model of Neuropathic Pain
To Be Published
|
|
1YF8
 
 | | Crystal structure of Himalayan mistletoe RIP reveals the presence of a natural inhibitor and a new functionally active sugar-binding site | | Descriptor: | 2-AMINO-4-ISOPROPYL-PTERIDINE-6-CARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mishra, V, Bilgrami, S, Sharma, R.S, Kaur, P, Yadav, S, Betzel, C, Babu, C.R, Singh, T.P. | | Deposit date: | 2004-12-31 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of himalayan mistletoe ribosome-inactivating protein reveals the presence of a natural inhibitor and a new functionally active sugar-binding site.
J.Biol.Chem., 280, 2005
|
|
8ZHF
 
 | | SARS-CoV-2 spike trimer (6P) in complex with R1-26 Fab, head-to-head aggregate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-26 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (5.26 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein helping to drive the genesis of Omicron variants
To Be Published
|
|
8ZHG
 
 | | SARS-CoV-2 spike trimer (6P) in complex with R1-26 Fab, focused refinement of RBD-Fab region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-26 Fab, Light chain of R1-26 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein helping to drive the genesis of Omicron variants
To Be Published
|
|
1I11
 
 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN, SOX-5 HMG BOX FROM MOUSE | | Descriptor: | TRANSCRIPTION FACTOR SOX-5 | | Authors: | Cary, P.D, Read, C.M, Davis, B, Driscoll, P.C, Crane-Robinson, C. | | Deposit date: | 2001-01-30 | | Release date: | 2001-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the DNA-binding domain of mouse Sox-5.
Protein Sci., 10, 2001
|
|
8ZHI
 
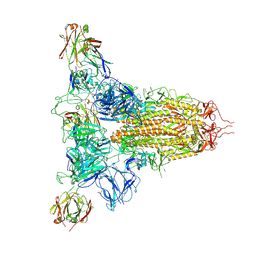 | | SARS-CoV-2 spike trimer (6P) in complex with three H18 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (6.05 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein helping to drive the genesis of Omicron variants
To Be Published
|
|
1PGJ
 
 | | X-RAY STRUCTURE OF 6-PHOSPHOGLUCONATE DEHYDROGENASE FROM THE PROTOZOAN PARASITE T. BRUCEI | | Descriptor: | 6-PHOSPHOGLUCONATE DEHYDROGENASE, SULFATE ION | | Authors: | Dohnalek, J, Phillips, C, Gover, S, Barrett, M.P, Adams, M.J. | | Deposit date: | 1998-03-16 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | A 2.8 A resolution structure of 6-phosphogluconate dehydrogenase from the protozoan parasite Trypanosoma brucei: comparison with the sheep enzyme accounts for differences in activity with coenzyme and substrate analogues.
J.Mol.Biol., 282, 1998
|
|
8ZHL
 
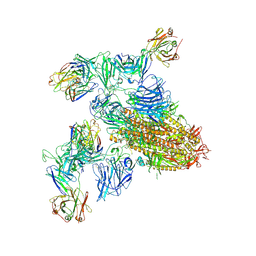 | | SARS-CoV-2 spike trimer (6P) in complex with two H18 and two R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein helping to drive the genesis of Omicron variants
To Be Published
|
|
8ZHM
 
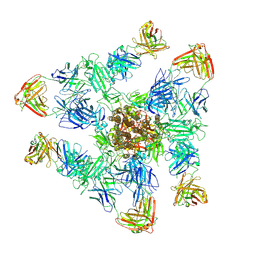 | | SARS-CoV-2 spike trimer (6P) in complex with three H18 and three R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein helping to drive the genesis of Omicron variants
To Be Published
|
|
5LIY
 
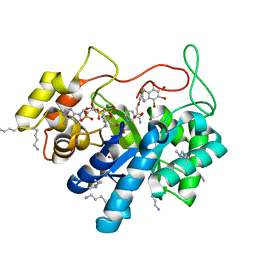 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor MK204 | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | Deposit date: | 2016-07-15 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
2W0Q
 
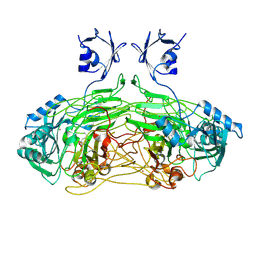 | | E. coli copper amine oxidase in complex with Xenon | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE, ... | | Authors: | Pirrat, P, Smith, M.A, Pearson, A.R, McPherson, M.J, Phillips, S.E.V. | | Deposit date: | 2008-08-20 | | Release date: | 2008-12-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of a Xenon Derivative of Escherichia Coli Copper Amine Oxidase: Confirmation of the Proposed Oxygen-Entry Pathway.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
4BR4
 
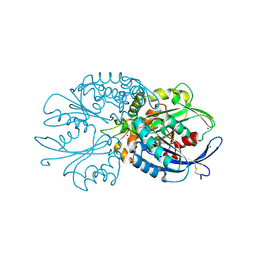 | | Legionella pneumophila NTPDase1 crystal form I, open, apo | | Descriptor: | CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, MAGNESIUM ION | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-03 | | Release date: | 2013-07-17 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
3RQU
 
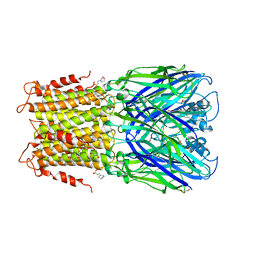 | | Crystal structure of a prokaryotic pentameric ligand-gated ion channel, ELIC | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ELIC Pentameric Ligand Gated Ion Channel from Erwinia Chrysanthemi, GLYCEROL | | Authors: | Pan, J.J, Chen, Q, Yoshida, K, Cohen, A, Kong, X.P, Xu, Y, Tang, P. | | Deposit date: | 2011-04-28 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.089 Å) | | Cite: | Structure of the pentameric ligand-gated ion channel ELIC cocrystallized with its competitive antagonist acetylcholine.
Nat Commun, 3, 2012
|
|
4BRM
 
 | | Sulfur SAD phasing of the Legionella pneumophila NTPDase1 - crystal form III (closed) in complex with sulfate | | Descriptor: | CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, SULFATE ION | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
7SXB
 
 | |
2VO7
 
 | | Structure of PKA complexed with 4-(4-Chlorobenzyl)-1-(7H-pyrrolo(2,3- d)pyrimidin-4-yl)piperidin-4-ylamine | | Descriptor: | 4-(4-chlorobenzyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-aminium, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Caldwell, J.J, Davies, T.G, Donald, A, McHardy, T, Rowlands, M.G, Aherne, G.W, Hunter, L.K, Taylor, K, Ruddle, R, Raynaud, F.I, Verdonk, M, Workman, P, Garrett, M.D, Collins, I. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Identification of 4-(4-Aminopiperidin-1-Yl)-7H-Pyrrolo[2,3-D]Pyrimidines as Selective Inhibitors of Protein Kinase B Through Fragment Elaboration.
J.Med.Chem., 51, 2008
|
|
5LL2
 
 | |
7QSA
 
 | |
4BQZ
 
 | | Rat NTPDase2 in complex with Mg GMPPNP | | Descriptor: | ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 2, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-03 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
2XXZ
 
 | | Crystal structure of the human JMJD3 jumonji domain | | Descriptor: | 1,2-ETHANEDIOL, 8-hydroxyquinoline-5-carboxylic acid, LYSINE-SPECIFIC DEMETHYLASE 6B, ... | | Authors: | Che, K.H, Yue, W.W, Krojer, T, Muniz, J.R.C, Ng, S.S, Tumber, A, Daniel, M, Burgess-Brown, N, Savitsky, P, Ugochukwu, E, Filippakopoulos, P, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2010-11-12 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Human Jmjd3 Jumonji Domain
To be Published
|
|
4BRD
 
 | | Legionella pneumophila NTPDase1 Q193E crystal form II, closed, Mg AMPPNP complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
2VU1
 
 | | Biosynthetic thiolase from Z. ramigera. Complex of with O-pantheteine- 11-pivalate. | | Descriptor: | ACETYL-COA ACETYLTRANSFERASE, PANTOTHENYL-AMINOETHANOL-11-PIVALIC ACID, SODIUM ION, ... | | Authors: | Kursula, P, Schmitz, W, Wierenga, R.K. | | Deposit date: | 2008-05-19 | | Release date: | 2008-10-28 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The Sulfur Atoms of the Substrate Coa and the Catalytic Cysteine are Required for a Productive Mode of Substrate Binding in Bacterial Biosynthetic Thiolase, a Thioester-Dependent Enzyme.
FEBS J., 275, 2008
|
|
