5L76
 
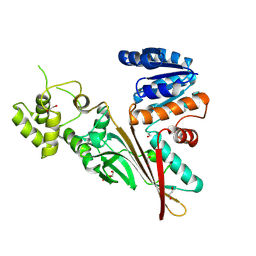 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain (in apo form) | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, mitochondrial, ... | | Authors: | Kopec, J, Pena, I.A, Rembeza, E, Strain-Damerell, C, Chalk, R, Borkowska, O, Goubin, S, Velupillai, S, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, Arruda, P, Yue, W.W. | | Deposit date: | 2016-06-02 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain (in apo form)
To Be Published
|
|
3RY6
 
 | | Complex of fcgammariia (CD32) and the FC of human IGG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ig gamma-1 chain C region, ... | | Authors: | Ramsland, P.A, Farrugia, W, Scott, A.M, Hogarth, P.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Basis for Fc{gamma}RIIa Recognition of Human IgG and Formation of Inflammatory Signaling Complexes.
J.Immunol., 187, 2011
|
|
5KZY
 
 | |
3RVF
 
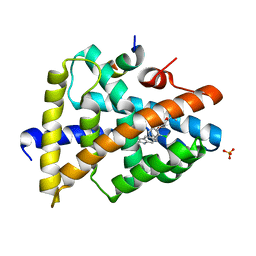 | | FXR with SRC1 and GSK2034 | | Descriptor: | 5-(4-{[3-(2,6-dichlorophenyl)-5-(propan-2-yl)-1,2-oxazol-4-yl]methoxy}phenyl)-1H-indole-2-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Williams, S.P, Madauss, K.P. | | Deposit date: | 2011-05-06 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformationally constrained farnesoid X receptor (FXR) agonists: Alternative replacements of the stilbene.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4CQA
 
 | | Plasmodium falciparum dihydroorotate dehydrogenase (DHODH) in complex with IDI-6273 | | Descriptor: | 2-(4-chlorobenzyl)-8-ethoxy-1,3-dimethylcyclohepta[c]pyrrol-4(2H)-one, DIHYDROOROTATE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Rowland, P. | | Deposit date: | 2014-02-12 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | In Vitro Resistance Selections for Plasmodium Falciparum Dihydroorotate Dehydrogenase Inhibitors Give Mutants with Multiple Point Mutations in the Drug-Binding Site and Altered Growth.
J.Biol.Chem., 289, 2014
|
|
2LWW
 
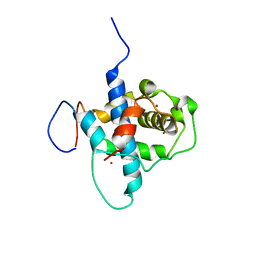 | | NMR structure of RelA-TAD/CBP-TAZ1 complex | | Descriptor: | CREB-binding protein, Nuclear transcription factor RelA, ZINC ION | | Authors: | Mukherjee, S.P, Ghosh, G, Wright, P.E. | | Deposit date: | 2012-08-07 | | Release date: | 2013-09-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Analysis of the RelA:CBP/p300 Interaction Reveals Its Involvement in NF-kappa B-Driven Transcription.
Plos Biol., 11, 2013
|
|
5KJO
 
 | |
1EXX
 
 | | ENANTIOMER DISCRIMINATION ILLUSTRATED BY CRYSTAL STRUCTURES OF THE HUMAN RETINOIC ACID RECEPTOR HRARGAMMA LIGAND BINDING DOMAIN: THE COMPLEX WITH THE INACTIVE S-ENANTIOMER BMS270395. | | Descriptor: | 3-FLUORO-4-[2-HYDROXY-2-(5,5,8,8-TETRAMETHYL-5,6,7,8,-TETRAHYDRO-NAPHTALEN-2-YL)-ACETYLAMINO]-BENZOIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-2 | | Authors: | Klaholz, B.P, Mitschler, A, Belema, M, Zusi, C, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-05-05 | | Release date: | 2000-06-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Enantiomer discrimination illustrated by high-resolution crystal structures of the human nuclear receptor hRARgamma.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
3NYN
 
 | | Crystal Structure of G Protein-Coupled Receptor Kinase 6 in Complex with Sangivamycin | | Descriptor: | (R,R)-2,3-BUTANEDIOL, G protein-coupled receptor kinase 6, SANGIVAMYCIN, ... | | Authors: | Tesmer, J.J.G, Singh, P. | | Deposit date: | 2010-07-15 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Molecular basis for activation of G protein-coupled receptor kinases.
Embo J., 29, 2010
|
|
1AZV
 
 | | FAMILIAL ALS MUTANT G37R CUZNSOD (HUMAN) | | Descriptor: | COPPER (II) ION, COPPER/ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Liu, H, Pellegrini, M, Nersissian, A.M, Gralla, E.B, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1997-11-21 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Subunit asymmetry in the three-dimensional structure of a human CuZnSOD mutant found in familial amyotrophic lateral sclerosis.
Protein Sci., 7, 1998
|
|
1B0Q
 
 | | DITHIOL ALPHA MELANOTROPIN PEPTIDE CYCLIZED VIA RHENIUM METAL COORDINATION | | Descriptor: | PROTEIN (CYCLIC ALPHA MELANOCYTE STIMULATING HORMONE), RHENIUM | | Authors: | Giblin, M.F, Wang, N, Hoffman, T.J, Jurisson, S.S, Quinn, T.P. | | Deposit date: | 1998-11-12 | | Release date: | 1998-11-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of alpha-melanotropin peptide analogs cyclized through rhenium and technetium metal coordination.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
5AOH
 
 | | Crystal Structure of CarF | | Descriptor: | POTASSIUM ION, Spore coat protein CotH | | Authors: | Tichy, E.M, Hardwick, S.W, Luisi, B.F, C Salmond, G.P. | | Deposit date: | 2015-09-10 | | Release date: | 2017-01-25 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 angstrom resolution crystal structure of the carbapenem intrinsic resistance protein CarF.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
8I2H
 
 | | Follicle stimulating hormone receptor | | Descriptor: | Follicle-stimulating hormone receptor | | Authors: | Duan, J, Xu, P, Yang, J, Ji, Y, Zhang, H, Mao, C, Luan, X, Jiang, Y, Zhang, Y, Zhang, S, Xu, H.E. | | Deposit date: | 2023-01-14 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Mechanism of hormone and allosteric agonist mediated activation of follicle stimulating hormone receptor.
Nat Commun, 14, 2023
|
|
3V7Z
 
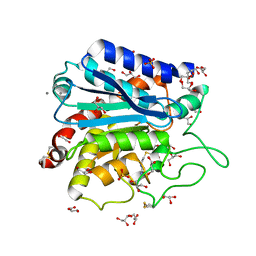 | | Carboxypeptidase T with GEMSA | | Descriptor: | (2-GUANIDINOETHYLMERCAPTO)SUCCINIC ACID, CALCIUM ION, Carboxypeptidase T, ... | | Authors: | Kuznetsov, S.A, Timofeev, V.I, Akparov, V.K, Kuranova, I.P. | | Deposit date: | 2011-12-22 | | Release date: | 2012-12-26 | | Last modified: | 2015-09-30 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into the broad substrate specificity of carboxypeptidase T from Thermoactinomyces vulgaris.
Febs J., 282, 2015
|
|
3OAB
 
 | | Mint deletion mutant of heterotetrameric geranyl pyrophosphate synthase in complex with ligands | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DIMETHYLALLYL S-THIOLODIPHOSPHATE, ... | | Authors: | Hsieh, F.-L, Chang, T.-H, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2010-08-05 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enhanced specificity of mint geranyl pyrophosphate synthase by modifying the R-loop interactions
J.Mol.Biol., 404, 2010
|
|
5KJ9
 
 | |
1B55
 
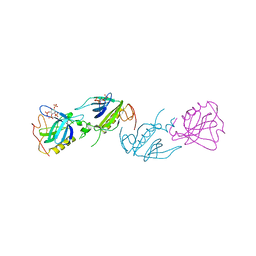 | | PH DOMAIN FROM BRUTON'S TYROSINE KINASE IN COMPLEX WITH INOSITOL 1,3,4,5-TETRAKISPHOSPHATE | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, TYROSINE-PROTEIN KINASE BTK, ZINC ION | | Authors: | Djinovic Carugo, K, Baraldi, E, Hyvoenen, M, Lo Surdo, P, Riley, A.M, Potter, B.V.L, O'Brien, R, Ladbury, J.E, Saraste, M. | | Deposit date: | 1999-01-12 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the PH domain from Bruton's tyrosine kinase in complex with inositol 1,3,4,5-tetrakisphosphate.
Structure Fold.Des., 7, 1999
|
|
3EUF
 
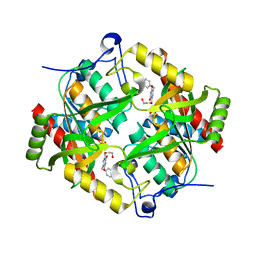 | | Crystal structure of BAU-bound human uridine phosphorylase 1 | | Descriptor: | 1-((2-HYDROXYETHOXY)METHYL)-5-BENZYLPYRIMIDINE-2,4(1H,3H)-DIONE, PHOSPHATE ION, Uridine phosphorylase 1 | | Authors: | Roosild, T.P. | | Deposit date: | 2008-10-10 | | Release date: | 2009-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Implications of the structure of human uridine phosphorylase 1 on the development of novel inhibitors for improving the therapeutic window of fluoropyrimidine chemotherapy.
Bmc Struct.Biol., 9, 2009
|
|
1IRL
 
 | | THE SOLUTION STRUCTURE OF THE F42A MUTANT OF HUMAN INTERLEUKIN 2 | | Descriptor: | INTERLEUKIN-2 | | Authors: | Mott, H.R, Baines, B.S, Hall, R.M, Cooke, R.M, Driscoll, P.C, Weir, M.P, Campbell, I.D. | | Deposit date: | 1995-08-25 | | Release date: | 1995-12-07 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the F42A mutant of human interleukin 2.
J.Mol.Biol., 247, 1995
|
|
5KKJ
 
 | | 2.0-Angstrom In situ Mylar structure of hen egg-white lysozyme (HEWL) at 293 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Broecker, J, Ernst, O.P. | | Deposit date: | 2016-06-21 | | Release date: | 2017-02-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | A Versatile System for High-Throughput In Situ X-ray Screening and Data Collection of Soluble and Membrane-Protein Crystals.
Cryst Growth Des, 16, 2016
|
|
4D9X
 
 | | The crystal structure of Coptisine bound to DNA d(CGTACG) | | Descriptor: | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium, CALCIUM ION, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Bilia, A.R. | | Deposit date: | 2012-01-12 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The crystal structure of Coptisine bound to DNA d(CGTACG)
to be published
|
|
1EN1
 
 | | STRUCTURE OF THE HIV-1 MINUS STRAND PRIMER BINDING SITE | | Descriptor: | DNA (5'-D(P*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3') | | Authors: | Johnson, P.E, Turner, R.B, Wu, Z.R, Levin, J.G, Summers, M.F. | | Deposit date: | 2000-03-20 | | Release date: | 2000-04-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription
Biochemistry, 39, 2000
|
|
4CVJ
 
 | | Neutron Structure of Compound I intermediate of Cytochrome c Peroxidase - Deuterium exchanged 100 K | | Descriptor: | CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Casadei, C.M, Gumiero, A, Blakeley, M.P, Ostermann, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (2.182 Å), X-RAY DIFFRACTION | | Cite: | Neutron Cryo-Crystallography Captures the Protonation State of Ferryl Heme in a Peroxidase
Science, 345, 2014
|
|
1IM1
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1, 20 STRUCTURES | | Descriptor: | ALPHA-CONOTOXIN IM1 | | Authors: | Rogers, J.P, Luginbuhl, P, Shen, G.S, Mccabe, R.T, Stevens, R.C, Wemmer, D.E. | | Deposit date: | 1998-11-18 | | Release date: | 1999-06-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-conotoxin ImI and comparison to other conotoxins specific for neuronal nicotinic acetylcholine receptors.
Biochemistry, 38, 1999
|
|
5KKW
 
 | |
