5EGL
 
 | | The structural and biochemical characterization of acyl-coa hydrolase from Staphylococcus aureus in complex with Butyryl Coenzyme A, Coenzyme A, and Coenzyme A disulfide | | Descriptor: | Acyl CoA Hydrolase, Butyryl Coenzyme A, COENZYME A, ... | | Authors: | Khandokar, Y.B, Srivastava, P.S, Forwood, J.K. | | Deposit date: | 2015-10-27 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural and biochemical characterization of acyl-coa hydrolase from Staphylococcus aureus
To Be Published
|
|
2W1E
 
 | | Structure determination of Aurora Kinase in complex with inhibitor | | Descriptor: | 4-[(2-{4-[(PHENYLCARBAMOYL)AMINO]-1H-PYRAZOL-3-YL}-1H-BENZIMIDAZOL-5-YL)METHYL]MORPHOLIN-4-IUM, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Howard, S, Berdini, V, Boulstridge, J.A, Carr, M.G, Cross, D.M, Curry, J, Devine, L.A, Early, T.R, Fazal, L, Gill, A.L, Heathcote, M, Maman, S, Matthews, J.E, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Rees, D.C, Reule, M, Tisi, D, Williams, G, Vinkovic, M, Wyatt, P.G. | | Deposit date: | 2008-10-17 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Fragment-Based Discovery of the Pyrazol-4-Yl Urea (at9283), a Multitargeted Kinase Inhibitor with Potent Aurora Kinase Activity.
J.Med.Chem., 52, 2009
|
|
2VO9
 
 | | CRYSTAL STRUCTURE OF THE ENZYMATICALLY ACTIVE DOMAIN OF THE LISTERIA MONOCYTOGENES BACTERIOPHAGE 500 ENDOLYSIN PLY500 | | Descriptor: | L-ALANYL-D-GLUTAMATE PEPTIDASE, SULFATE ION, ZINC ION | | Authors: | Korndoerfer, I.P, Kanitz, A, Skerra, A. | | Deposit date: | 2008-02-09 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of the L-Alanoyl-D-Glutamate Endopeptidase Domain of Listeria Bacteriophage Endolysin Ply500 Reveals a New Member of the Las Peptidase Family.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
5EJU
 
 | |
2W7R
 
 | | Structure of the PDZ domain of Human Microtubule associated serine- threonine kinase 4 | | Descriptor: | MICROTUBULE-ASSOCIATED SERINE/THREONINE-PROTEIN KINASE 4, PHOSPHATE ION | | Authors: | Muniz, J.R.C, Elkins, J, Wang, J, Savitzky, P, Roos, A, Salah, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S. | | Deposit date: | 2008-12-24 | | Release date: | 2009-05-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unusual Binding Interactions in Pdz Domain Crystal Structures Help Explain Binding Mechanisms.
Protein Sci., 19, 2010
|
|
5EKK
 
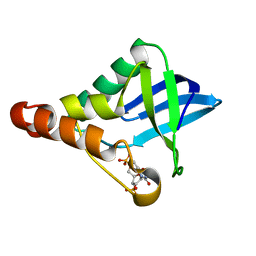 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V39D/L125E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Skerritt, L.A, Bell-Upp, P.C, Siegler, M.A, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-11-03 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V39D/L125E at cryogenic temperature
To be Published
|
|
2VS4
 
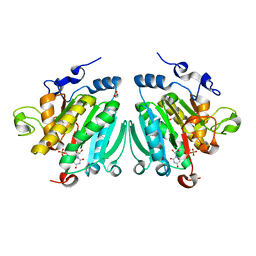 | | THE BINDING OF UDP-GALACTOSE BY AN ACTIVE SITE MUTANT OF alpha-1,3 GALACTOSYLTRANSFERASE (alpha3GT) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Brew, K, Acharya, K.R. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Basis of Udp-Galactose Binding by Alpha- 1,3-Galactosyltransferase (Alpha3Gt): Role of Negative Charge on Aspartic Acid 316 in Structure and Activity.
Biochemistry, 47, 2008
|
|
2W7W
 
 | |
2WA7
 
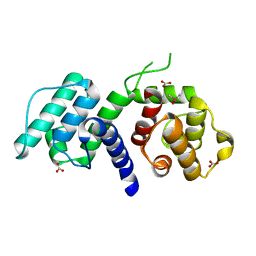 | | Structure of the M202V mutant of human filamin b actin binding domain at 1.85 Angstrom resolution | | Descriptor: | CACODYLATE ION, CARBONATE ION, FILAMIN-B | | Authors: | Sawyer, G.M, Clark, A.R, Robertson, S.P, Sutherland-Smith, A.J. | | Deposit date: | 2009-02-03 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Disease-Associated Substitutions in the Filamin B Actin Binding Domain Confer Enhanced Actin Binding Affinity in the Absence of Major Structural Disturbance: Insights from the Crystal Structures of Filamin B Actin Binding Domains.
J.Mol.Biol., 390, 2009
|
|
2W4B
 
 | | Epstein-Barr virus alkaline nuclease D203S mutant | | Descriptor: | ALKALINE EXONUCLEASE | | Authors: | Buisson, M, Geoui, T, Flot, D, Tarbouriech, N, Burmeister, W.P. | | Deposit date: | 2008-11-24 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A Bridge Crosses the Active Site Canyon of the Epstein-Barr Virus Nuclease with DNase and Rnase Activity.
J.Mol.Biol., 391, 2009
|
|
5EOK
 
 | | Human Plasma Coagulation Factor XI in complex with peptide P39 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XI, ... | | Authors: | Wong, S.S, Ostergaard, S, Hall, G, Li, C, Williams, P.M, Stennicke, H, Emsley, J. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A novel DFP tripeptide motif interacts with the coagulation factor XI apple 2 domain.
Blood, 127, 2016
|
|
2W1I
 
 | | Structure determination of Aurora Kinase in complex with inhibitor | | Descriptor: | 4-[(2-{4-[(CYCLOPROPYLCARBAMOYL)AMINO]-1H-PYRAZOL-3-YL}-1H-BENZIMIDAZOL-6-YL)METHYL]MORPHOLIN-4-IUM, JAK2 | | Authors: | Howard, S, Berdini, V, Boulstridge, J.A, Carr, M.G, Cross, D.M, Curry, J, Devine, L.A, Early, T.R, Fazal, L, Gill, A.L, Heathcote, M, Maman, S, Matthews, J.E, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Rees, D.C, Reule, M, Tisi, D, Williams, G, Vinkovic, M, Wyatt, P.G. | | Deposit date: | 2008-10-17 | | Release date: | 2009-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fragment-Based Discovery of the Pyrazol-4-Yl Urea (at9283), a Multitargeted Kinase Inhibitor with Potent Aurora Kinase Activity.
J.Med.Chem., 52, 2009
|
|
2WAS
 
 | |
5EPM
 
 | |
6TT2
 
 | |
2W5N
 
 | | Native structure of the GH93 alpha-L-arabinofuranosidase of Fusarium graminearum | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-L-ARABINOFURANOSIDASE, GLYCEROL, ... | | Authors: | Carapito, R, Imberty, A, Jeltsch, J.M, Byrns, S.C, Tam, P.H, Lowary, T.L, Varrot, A, Phalip, V. | | Deposit date: | 2008-12-11 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Basis of Arabinobio-Hydrolase Activity in Phytopathogenic Fungi. Crystal Structure and Catalytic Mechanism of Fusarium Graminearum Gh93 Exo-Alpha-L-Arabinanase.
J.Biol.Chem., 284, 2009
|
|
2WGV
 
 | | Crystal structure of the OXA-10 V117T mutant at pH 6.5 inhibited by a chloride ion | | Descriptor: | BETA-LACTAMASE OXA-10, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Vercheval, L, Kerff, F, Bauvois, C, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | Deposit date: | 2009-04-27 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post- Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
5EGK
 
 | |
2WB3
 
 | | The partial structure of a group A streptococcal phage-encoded tail fibre hyaluronate lyase Hylp3 | | Descriptor: | HYALURONIDASE-PHAGE ASSOCIATED | | Authors: | Martinez-Fleites, C, Black, G.W, Turkenburg, J.P, Smith, N.L, Taylor, E.J. | | Deposit date: | 2009-02-20 | | Release date: | 2009-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Two Truncated Phage-Tail Hyaluronate Lyases from Streptococcus Pyogenes Serotype M1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2WBT
 
 | | The Structure of a Double C2H2 Zinc Finger Protein from a Hyperthermophilic Archaeal Virus in the Absence of DNA | | Descriptor: | B-129, ZINC ION | | Authors: | Eilers, B.J, Menon, S, Windham, A.B, Kraft, P, Dlakic, M, Young, M.J, Lawrence, C.M. | | Deposit date: | 2009-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of a Double C2H2 Zinc Finger Protein from a Hyperthermophilic Archaeal Virus in the Absence of DNA
To be Published
|
|
5EIJ
 
 | | Carbonic Anhydrase II in complex with Sulfonamide Inhibitor | | Descriptor: | 1-(3-iodanylphenyl)-3-(4-sulfamoylphenyl)thiourea, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Lomelino, C.L, Mahon, B.P, McKenna, R. | | Deposit date: | 2015-10-29 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Kinetic and X-ray crystallographic investigations on carbonic anhydrase isoforms I, II, IX and XII of a thioureido analog of SLC-0111.
Bioorg. Med. Chem., 24, 2016
|
|
2WCO
 
 | | Structures of the Streptomyces coelicolor A3(2) Hyaluronan Lyase in Complex with Oligosaccharide Substrates and an Inhibitor | | Descriptor: | 4-deoxy-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, FORMIC ACID, GLYCEROL, ... | | Authors: | Elmabrouk, Z.H, Taylor, E.J, Vincent, F, Smith, N.L, Zhang, M, Charnock, S.J, Turkenburg, J.P, Davies, G.J, Black, G.W. | | Deposit date: | 2009-03-12 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structures of a Family 8 Polysaccharide Lyase Reveal Open and Highly Occluded Substrate-Binding Cleft Conformations.
Proteins, 79, 2011
|
|
5ENN
 
 | | The crystal structure of Human VPS34 in complex with a selective and potent inhibitor | | Descriptor: | 1-[[4-(cyclopropylmethyl)-5-[2-(pyridin-4-ylamino)pyrimidin-4-yl]pyrimidin-2-yl]amino]-2-methyl-propan-2-ol, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3, ... | | Authors: | Kearney, E.P. | | Deposit date: | 2015-11-09 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Potent, selective, and orally bioavailable inhibitors of VPS34 provide chemical tools to modulate autophagy in vivo
To Be Published
|
|
6T1N
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 5 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
5EP1
 
 | | Quorum-Sensing Signal Integrator LuxO - Catalytic Domain | | Descriptor: | ACETATE ION, Putative repressor protein luxO | | Authors: | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2015-11-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
