6YTC
 
 | |
4BTT
 
 | | factor Xa in complex with the dual thrombin-FXa inhibitor 31. | | Descriptor: | CALCIUM ION, COAGULATION FACTOR X, COAGULATION FACTOR X LIGHT CHAIN, ... | | Authors: | Meneyrol, J, Follmann, M, Lassalle, G, Wehner, V, Barre, G, Rousseaux, T, Altenburger, J.M, Petit, F, Bocskei, Z, Stehlin-Gaon, C, Schreuder, H, Alet, N, Herault, J.-P, Millet, L, Dol, F, Hasbrand, C, Schaeffer, P, Sadoun, F, Klieber, S, Briot, C, Bono, F, Herbert, J.-M. | | Deposit date: | 2013-06-19 | | Release date: | 2013-12-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | 5-Chlorothiophene-2-Carboxylic Acid [(S)-2-[2-Methyl-3-(2-Oxopyrrolidin-1-Yl)Benzenesulfonylamino]-3-(4-Methylpiperazin-1-Yl)-3-Oxopropyl]Amide (Sar107375), a Selective and Potent Orally Active Dual Thrombin and Factor Xa Inhibitor.
J.Med.Chem., 56, 2013
|
|
7D76
 
 | | Cryo-EM structure of the beclomethasone-bound adhesion receptor GPR97-Go complex | | Descriptor: | (8~{S},9~{R},10~{S},11~{S},13~{S},14~{S},16~{S},17~{R})-9-chloranyl-10,13,16-trimethyl-11,17-bis(oxidanyl)-17-(2-oxidanylethanoyl)-6,7,8,11,12,14,15,16-octahydrocyclopenta[a]phenanthren-3-one, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | Authors: | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
1M0V
 
 | | NMR STRUCTURE OF THE TYPE III SECRETORY DOMAIN OF YERSINIA YOPH COMPLEXED WITH THE SKAP-HOM PHOSPHO-PEPTIDE N-acetyl-DEpYDDPF-NH2 | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YOPH, SKAP55 homologue | | Authors: | Khandelwal, P, Keliikuli, K, Smith, C.L, Saper, M.A, Zuiderweg, E.R.P. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and phosphopeptide binding to the N-terminal domain of Yersinia YopH: comparison with a crystal structure
Biochemistry, 41, 2002
|
|
4BTU
 
 | | Factor Xa in complex with the dual thrombin-FXa inhibitor 57. | | Descriptor: | 5-Chloro-thiophene-2-carboxylic acid [(S)-2-[2-chloro-5-fluoro-3-(2-oxo-piperidin-1-yl)-benzenesulfonylamino]-3-(4-methyl-piperazin-1-yl)-3-oxo-propyl]-amide, CALCIUM ION, COAGULATION FACTOR X HEAVY CHAIN, ... | | Authors: | Meneyrol, J, Follmann, M, Lassalle, G, Wehner, V, Barre, G, Rousseaux, T, Altenburger, J.M, Petit, F, Bocskei, Z, Stehlin-Gaon, C, Schreuder, H, Alet, N, Herault, J.-P, Millet, L, Dol, F, Hasbrand, C, Schaeffer, P, Sadoun, F, Klieber, S, Briot, C, Bono, F, Herbert, J.-M. | | Deposit date: | 2013-06-19 | | Release date: | 2013-12-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | 5-Chlorothiophene-2-Carboxylic Acid [(S)-2-[2-Methyl-3-(2-Oxopyrrolidin-1-Yl)Benzenesulfonylamino]-3-(4-Methylpiperazin-1-Yl)-3-Oxopropyl]Amide (Sar107375), a Selective and Potent Orally Active Dual Thrombin and Factor Xa Inhibitor.
J.Med.Chem., 56, 2013
|
|
7RZ6
 
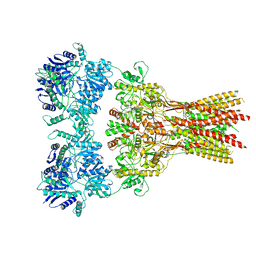 | | Structure of the complex of AMPA receptor GluA2 with auxiliary subunit TARP gamma-5 bound to agonist glutamate | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Klykov, O.V, Gangwar, S.P, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
7RYY
 
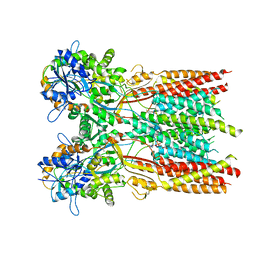 | | Structure of the complex of LBD-TMD part of AMPA receptor GluA2 with auxiliary subunit TARP gamma-5 bound to agonist glutamate | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Klykov, O.V, Gangwar, S.P, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-26 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
5X00
 
 | |
1X07
 
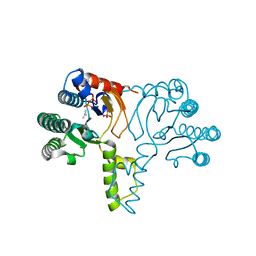 | | Crystal structure of undecaprenyl pyrophosphate synthase in complex with Mg and IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Guo, R.-T, Ko, T.-P, Chen, A.P.-C, Kuo, C.-J, Wang, A.H.-J, Liang, P.-H. | | Deposit date: | 2005-03-15 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of undecaprenyl pyrophosphate synthase in complex with magnesium, isopentenyl pyrophosphate, and farnesyl thiopyrophosphate: roles of the metal ion and conserved residues in catalysis.
J.Biol.Chem., 280, 2005
|
|
6R5L
 
 | | Fragment AZ-006 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 4-[[(2~{S})-1-azanylpropan-2-yl]amino]-6-(sulfanylmethyl)-1-benzothiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.884 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6DHR
 
 | |
7AAD
 
 | | Crystal structure of the catalytic domain of human PARP1 in complex with olaparib | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
1S74
 
 | | SOLUTION STRUCTURE OF A DNA DUPLEX CONTAINING AN ALPHA-ANOMERIC ADENOSINE: INSIGHTS INTO SUBSTRATE RECOGNITION BY ENDONUCLEASE IV | | Descriptor: | 5'-D(*CP*GP*TP*CP*GP*TP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*(A3A)P*CP*GP*AP*CP*G)-3' | | Authors: | Aramini, J.M, Cleaver, S.H, Pon, R.T, Cunningham, R.P, Germann, M.W. | | Deposit date: | 2004-01-28 | | Release date: | 2004-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Duplex Containing an alpha-Anomeric Adenosine: Insights into Substrate Recognition by Endonuclease IV.
J.Mol.Biol., 338, 2004
|
|
7DHR
 
 | | Cryo-EM structure of the full agonist isoprenaline-bound beta2 adrenergic receptor-Gs protein complex. | | Descriptor: | Beta-2 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, F, Ling, S.L, Zhou, Y.X, Zhang, Y.N, Lv, P, Liu, S.L, Fang, W, Sun, W.J, Hu, L.Y.A. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Different Conformational Responses of the beta2-Adrenergic Receptor-Gs Complex upon Binding of the Partial Agonist Salbutamol or the Full Agonist Isoprenaline
Natl Sci Rev, 2020
|
|
6R7H
 
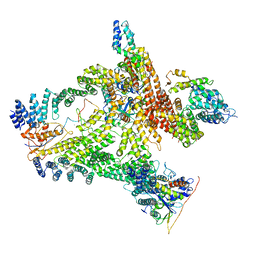 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
7A7N
 
 | |
7SF6
 
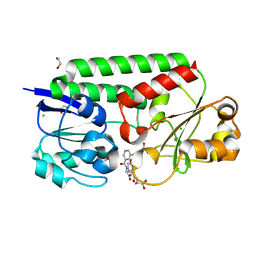 | | Crystal Structure of Siderophore Binding Protein FatB from Desulfitobacterium hafniense | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,3-DIHYDROXY-BENZOYLAMINO)-3-HYDROXY-PROPIONIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Patel, H.P, Nordquist, K.A, Schaab, K.M, Sha, J, Babnigg, G, Bond, A.H, Joachimiak, A, Midwest Center for Structural Genomics, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2021-10-03 | | Release date: | 2021-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of Siderophore Binding Protein FatB from Desulfitobacterium hafniense
To Be Published
|
|
4KAP
 
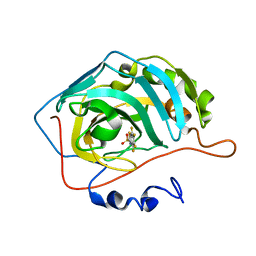 | | The Binding of Benzoarylsulfonamide Ligands to Human Carbonic Anhydrase is Insensitive to Formal Fluorination of the Ligand | | Descriptor: | 4,5,6,7-tetrafluoro-1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Lange, H, Lockett, M.R, Breiten, B, Heroux, A, Sherman, W, Rappoport, D, Yau, P.O, Snyder, P.W, Whitesides, G.M. | | Deposit date: | 2013-04-22 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Binding of Benzoarylsulfonamide Ligands to Human Carbonic Anhydrase is Insensitive to Formal Fluorination of the Ligand.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
7A7L
 
 | | rsEGFP in the green-off state | | Descriptor: | Green fluorescent protein, TETRAETHYLENE GLYCOL | | Authors: | De Zitter, E, Dedecker, P, Van Meervelt, L. | | Deposit date: | 2020-08-30 | | Release date: | 2021-02-17 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-Function Dataset Reveals Environment Effects within a Fluorescent Protein Model System*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7A7K
 
 | | rsEGFP in the green-on state | | Descriptor: | Green fluorescent protein, SULFATE ION | | Authors: | De Zitter, E, Dedecker, P, Van Meervelt, L. | | Deposit date: | 2020-08-30 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Function Dataset Reveals Environment Effects within a Fluorescent Protein Model System*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7A7T
 
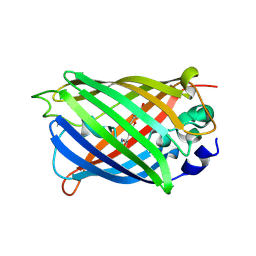 | |
7A7P
 
 | | rsGreen0.7-K206A partially in the green-off state | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, NITRATE ION | | Authors: | De Zitter, E, Dedecker, P, Van Meervelt, L. | | Deposit date: | 2020-08-30 | | Release date: | 2021-02-17 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure-Function Dataset Reveals Environment Effects within a Fluorescent Protein Model System*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7A7W
 
 | |
3VCR
 
 | | Crystal structure of a putative Kdpg (2-keto-3-deoxy-6-phosphogluconate) aldolase from Oleispira antarctica | | Descriptor: | PYRUVIC ACID, putative Kdpg (2-keto-3-deoxy-6-phosphogluconate) aldolase | | Authors: | Stogios, P.J, Kagan, O, Di Leo, R, Yim, V, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-04 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
1SD5
 
 | | Crystal structure of Rv1626 | | Descriptor: | IODIDE ION, putative antiterminator | | Authors: | Morth, J.P, Feng, V, Perry, L.J, Svergun, D.I, Tucker, P.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-02-13 | | Release date: | 2004-09-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Crystal and Solution Structure of a Putative Transcriptional Antiterminator from Mycobacterium tuberculosis.
Structure, 12, 2004
|
|
