5KSN
 
 | |
8GCW
 
 | | Stx2A1(Y77A)-P6 peptide | | Descriptor: | P stalk protein, rRNA N-glycosylase | | Authors: | Rudolph, M.J, Li, X.P. | | Deposit date: | 2023-03-03 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Stx2A1(Y77A) in complex with P6 peptide.
To Be Published
|
|
5E5A
 
 | | Crystal structure of the chromatin-tethering domain of Human cytomegalovirus IE1 protein bound to the nucleosome core particle | | Descriptor: | C-terminal domain of Regulatory protein IE1, DNA (146-MER), Histone H2A, ... | | Authors: | Fang, Q, Chen, P, Wang, M, Fang, J, Yang, N, Li, G, Xu, R.M. | | Deposit date: | 2015-10-08 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Human cytomegalovirus IE1 protein alters the higher-order chromatin structure by targeting the acidic patch of the nucleosome
Elife, 5, 2016
|
|
1AG3
 
 | |
5KUX
 
 | |
5L8Q
 
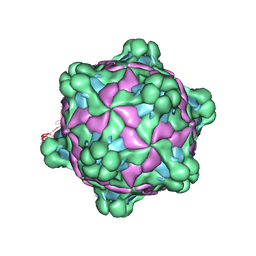 | | Structure of deformed wing virus, a honeybee pathogen | | Descriptor: | URIDINE-5'-MONOPHOSPHATE, VP1, VP2, ... | | Authors: | Skubnik, K, Novacek, J, Fuzik, T, Pridal, A, Paxton, R, Plevka, P. | | Deposit date: | 2016-06-08 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of deformed wing virus, a major honey bee pathogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7ZDQ
 
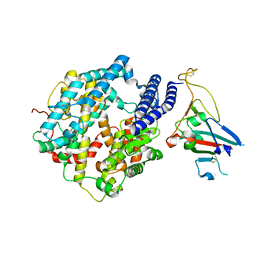 | | Cryo-EM structure of Human ACE2 bound to a high-affinity SARS CoV-2 mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Bate, N, Savva, C.G, Moody, P.C.E, Brown, E.A, Schwabe, W.R, Brindle, N.P.J, Ball, J.K, Sale, J.E. | | Deposit date: | 2022-03-29 | | Release date: | 2022-05-18 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | In vitro evolution predicts emerging SARS-CoV-2 mutations with high affinity for ACE2 and cross-species binding.
Plos Pathog., 18, 2022
|
|
7MJ2
 
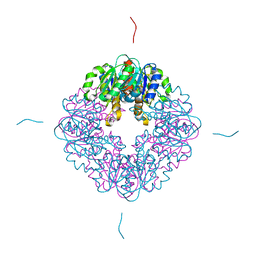 | | LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with Zn | | Descriptor: | MAGNESIUM ION, Pyridinium-3,5-biscarboxylic acid mononucleotide synthase, ZINC ION | | Authors: | Chatterjee, S, Rankin, J.A, Lagishetty, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2021-04-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The LarB carboxylase/hydrolase forms a transient cysteinyl-pyridine intermediate during nickel-pincer nucleotide cofactor biosynthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6DZI
 
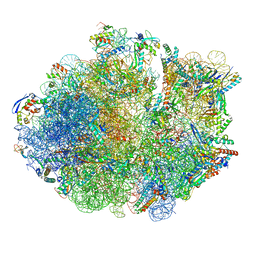 | | Cryo-EM Structure of Mycobacterium smegmatis 70S C(minus) ribosome 70S-MPY complex | | Descriptor: | 16S rRNA, 23 S rRNA (3119-MER), 30S ribosomal protein S10, ... | | Authors: | Sharma, M.R, Li, Y, Korripella, R, Yang, Y, Kaushal, P.S, Lin, Q, Wade, J.T, Gray, A.G, Derbyshire, K.M, Agrawal, R.K, Ojha, A. | | Deposit date: | 2018-07-05 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Zinc depletion induces ribosome hibernation in mycobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5E79
 
 | | Macromolecular diffractive imaging using imperfect crystals | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ayyer, K, Yefanov, O, Oberthur, D, Roy-Chowdhury, S, Galli, L, Mariani, V, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Schaffer, A, Dorner, K, James, D, Kupitz, C, Metz, M, Nelson, G, Xavier, P.L, Beyerlein, K.R, Schmidt, M, Sarrou, I, Spence, J.C.H, Weierstall, U, White, T.A, Yang, J.-H, Zhao, Y, Liang, M, Aquila, A, Hunter, M.S, Koglin, J.E, Boutet, S, Fromme, P, Barty, A, Chapman, H.N. | | Deposit date: | 2015-10-12 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Macromolecular diffractive imaging using imperfect crystals.
Nature, 530, 2016
|
|
8OKX
 
 | | Structure of cGAS in complex with SPSB3-ELOBC | | Descriptor: | Cyclic GMP-AMP synthase, Elongin-B, Elongin-C, ... | | Authors: | Xu, P.B, Ablasser, A. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | The CRL5-SPSB3 ubiquitin ligase targets nuclear cGAS for degradation.
Nature, 627, 2024
|
|
3HZG
 
 | | Crystal structure of mycobacterium tuberculosis thymidylate synthase X bound with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Staker, B.L, Rathod, P, Hunter, J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Increasing the structural coverage of tuberculosis drug targets.
Tuberculosis (Edinb), 95, 2015
|
|
5KQ2
 
 | |
5EA8
 
 | | Crystal Structure of Prefusion RSV F Glycoprotein Fusion Inhibitor Resistance Mutant D489Y | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, D(-)-TARTARIC ACID, Fusion glycoprotein F0, ... | | Authors: | Battles, M.B, McLellan, J.S, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
5KQI
 
 | |
5ZTJ
 
 | | Crystal Structure of GyraseA C-Terminal Domain from Salmonella typhi at 2.4A Resolution | | Descriptor: | DNA gyrase subunit A | | Authors: | Sachdeva, E, Gupta, D, Tiwari, P, Kaur, G, Sharma, S, Singh, T.P, Ethayathulla, A.S, Kaur, P. | | Deposit date: | 2018-05-03 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The pivot point arginines identified in the beta-pinwheel structure of C-terminal domain from Salmonella Typhi DNA Gyrase A subunit.
Sci Rep, 10, 2020
|
|
4WVG
 
 | |
5KSG
 
 | |
5KT0
 
 | | Dihydrodipicolinate reductase from the industrial and evolutionarily important cyanobacteria Anabaena variabilis. | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate reductase, MAGNESIUM ION | | Authors: | Christensen, J.B, Soares da Costa, T.P, Faou, P, Pearce, F.G, Panjikar, S, Perugini, M.A. | | Deposit date: | 2016-07-10 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Dihydrodipicolinate Reductase from the industrial and evolutionarily important cyanobacteria Anabaena variabilis.
To Be Published
|
|
5E0A
 
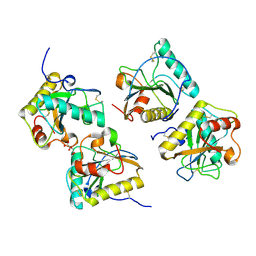 | | Crystal Structure of the complex of Camel Peptidoglycan Recognition Protein (CPGRP-S) and N-Acetylglucosamine at 2.6 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the complex of Camel Peptidoglycan Recognition Protein (CPGRP-S) and N-Acetylglucosamine at 2.6 A
To Be Published
|
|
5HPT
 
 | |
6RMR
 
 | |
5KU1
 
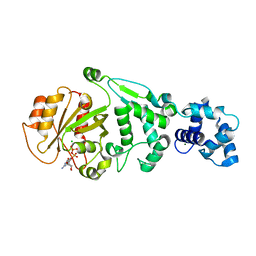 | | hMiro1 EF hand and cGTPase domains in the GDP-bound state | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
8OE3
 
 | |
8OE8
 
 | |
