4QGD
 
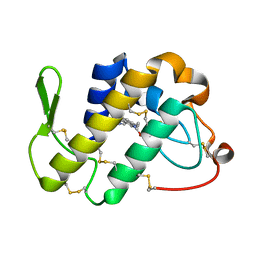 | | Crystal Structure of the Complex of Phospholipase A2 with Gramine derivative at 1.80 A Resolution | | Descriptor: | 3-{3-[(DIMETHYLAMINO)METHYL]-1H-INDOL-7-YL}PROPAN-1-OL, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Shukla, P.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-05-22 | | Release date: | 2014-06-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures and binding studies of the complexes of phospholipase A2 with five inhibitors
Biochim.Biophys.Acta, 1854, 2015
|
|
2P58
 
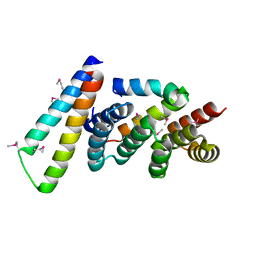 | | Structure of the Yersinia pestis Type III secretion system needle protein YscF in complex with its chaperones YscE/YscG | | Descriptor: | Putative type III secretion protein YscE, Putative type III secretion protein YscF, Putative type III secretion protein YscG | | Authors: | Sun, P, Austin, B.P, Tropea, J.E, Waugh, D.S. | | Deposit date: | 2007-03-14 | | Release date: | 2008-03-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the Yersinia pestis type III secretion system needle protein YscF in complex with its heterodimeric chaperone YscE/YscG.
J.Mol.Biol., 377, 2008
|
|
3DMJ
 
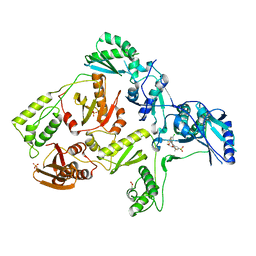 | | CRYSTAL STRUCTURE OF HIV-1 V106A and Y181C MUTANT REVERSE TRANSCRIPTASE IN COMPLEX WITH GW564511. | | Descriptor: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2008-07-01 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
3HY0
 
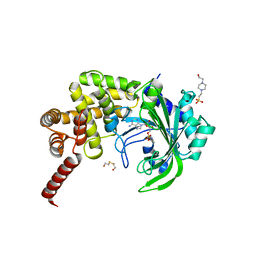 | | Crystal Structure of catalytic fragment of E. coli AlaRS G237A in complex with GlySA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-(glycylsulfamoyl)adenosine, ... | | Authors: | Guo, M, Yang, X.-L, Schimmel, P. | | Deposit date: | 2009-06-22 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma.
Nature, 462, 2009
|
|
4DG6
 
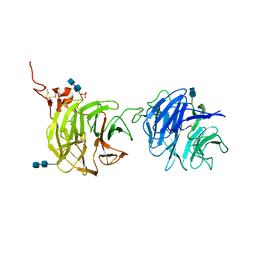 | | Crystal structure of domains 1 and 2 of LRP6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Low-density lipoprotein receptor-related protein 6, PHOSPHATE ION | | Authors: | Ceska, T.A, Doyle, C, Slocombe, P. | | Deposit date: | 2012-01-25 | | Release date: | 2012-06-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Characterization of the interaction of sclerostin with the LRP family of Wnt co-receptors
To be Published
|
|
2RDD
 
 | | X-ray crystal structure of AcrB in complex with a novel transmembrane helix. | | Descriptor: | (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, Acriflavine resistance protein B, UPF0092 membrane protein yajC | | Authors: | Tornroth-Horsefield, S, Gourdon, P, Horsefield, R, Neutze, R. | | Deposit date: | 2007-09-22 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of AcrB in complex with a single transmembrane subunit reveals another twist.
Structure, 15, 2007
|
|
1KJ5
 
 | | Solution Structure of Human beta-defensin 1 | | Descriptor: | BETA-DEFENSIN 1 | | Authors: | Schibli, D.J, Hunter, H.N, Aseyev, V, Starner, T.D, Wiencek, J.M, McCray Jr, P.B, Tack, B.F, Vogel, H.J. | | Deposit date: | 2001-12-04 | | Release date: | 2002-03-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the human beta-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus.
J.Biol.Chem., 277, 2002
|
|
3HZ8
 
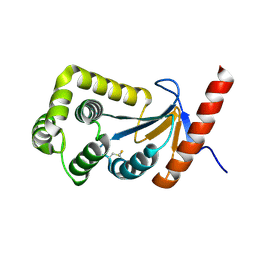 | | Crystal structure of the oxidized T176V DsbA1 mutant | | Descriptor: | Thiol:disulfide interchange protein DsbA | | Authors: | Lafaye, C, Iwema, T, Carpentier, P, Jullian-Binard, C, Serre, L. | | Deposit date: | 2009-06-23 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Biochemical and structural study of the homologues of the thiol-disulfide oxidoreductase DsbA in Neisseria meningitidis.
J.Mol.Biol., 392, 2009
|
|
4DGJ
 
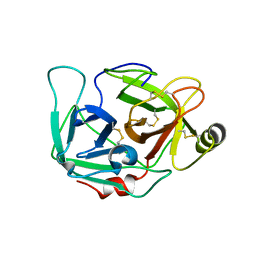 | |
5SXX
 
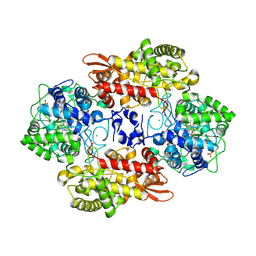 | |
4DHN
 
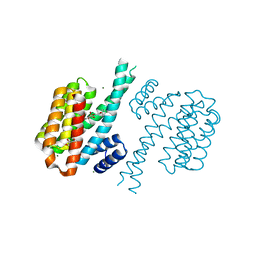 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
1NF4
 
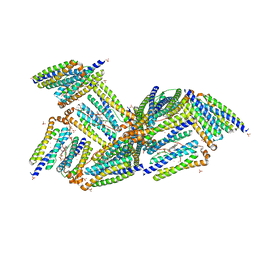 | | X-Ray Structure of the Desulfovibrio desulfuricans bacterioferritin: the diiron site in different states (reduced structure) | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, FE (II) ION, SULFATE ION, ... | | Authors: | Macedo, S, Romao, C.V, Mitchell, E, Matias, P.M, Liu, M.Y, Xavier, A.V, LeGall, J, Teixeira, M, Lindley, P, Carrondo, M.A. | | Deposit date: | 2002-12-13 | | Release date: | 2003-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The nature of the di-iron site in the bacterioferritin from
Desulfovibrio desulfuricans
NAT.STRUCT.BIOL., 10, 2003
|
|
1AGH
 
 | |
5SYU
 
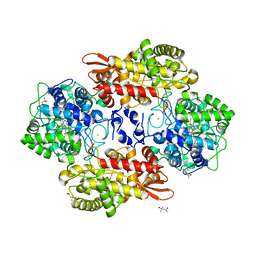 | | Crystal structure of Burkholderia pseudomallei KatG E242Q variant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Catalase-peroxidase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2016-08-12 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Catalase Activity of Catalase-Peroxidases Is Modulated by Changes in the pKa of the Distal Histidine.
Biochemistry, 56, 2017
|
|
1AML
 
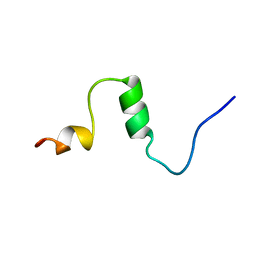 | |
1KG2
 
 | | Crystal structure of the core fragment of MutY from E.coli at 1.2A resolution | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1KL7
 
 | | Crystal Structure of Threonine Synthase from Yeast | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Threonine Synthase | | Authors: | Garrido-Franco, M, Ehlert, S, Messerschmidt, A, Marinkovic, S, Huber, R, Laber, B, Bourenkov, G.P, Clausen, T. | | Deposit date: | 2001-12-11 | | Release date: | 2002-04-24 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of threonine synthase from yeast.
J.Biol.Chem., 277, 2002
|
|
1KG7
 
 | | Crystal Structure of the E161A mutant of E.coli MutY (core fragment) | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
6B0I
 
 | | Apo KLP10A in complex with a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein Klp10A, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Sosa, H. | | Deposit date: | 2017-09-14 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Cryo-EM reveals the structural basis of microtubule depolymerization by kinesin-13s.
Nat Commun, 9, 2018
|
|
4HZE
 
 | | Crystal structure of human Arginase-2 complexed with inhibitor 9 | | Descriptor: | Arginase-2, mitochondrial, BENZAMIDINE, ... | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D.L, Golebiowski, A, Ji, M, Zhang, M, Beckett, P, Sheeler, R, Andreoli, M, Conway, B, Mahboubi, K, Schroeter, H, Van Zandt, M.C, Podjarny, A. | | Deposit date: | 2012-11-15 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Discovery of (R)-2-Amino-6-borono-2-(2-(piperidin-1-yl)ethyl)hexanoic Acid and Congeners As Highly Potent Inhibitors of Human Arginases I and II for Treatment of Myocardial Reperfusion Injury.
J.Med.Chem., 56, 2013
|
|
3HXX
 
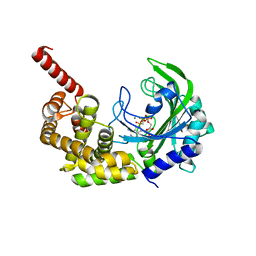 | |
1SL3
 
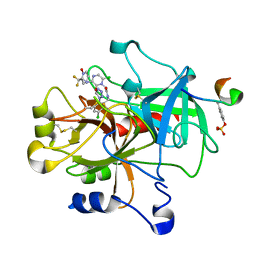 | | crystal structue of Thrombin in complex with a potent P1 heterocycle-Aryl based inhibitor | | Descriptor: | (2-[6-CHLORO-3-{[2,2-DIFLUORO-2-(1-OXIDOPYRIDIN-2-YL)ETHYL]AMINO}-2-OXOPYRAZIN-1(2H)-YL]-N-[5-CHLORO-2-(1H-TETRAZOL-1-YL)BENZYL]ACETAMIDE, Hirudin, thrombin | | Authors: | Young, M.B, Barrow, J.C, Glass, K.L, Lundell, G.F, Newton, C.L, Pellicore, J.M, Rittle, K.E, Selnick, H.G, Stauffer, K.J, Vacca, J.P, Williams, P.D, Bohn, D, Clayton, F.C, Cook, J.J, Krueger, J.A, Kuo, L.C, Lewis, S.D, Lucas, B.J, McMasters, D.R, Miller-Stein, C, Pietrak, B.L. | | Deposit date: | 2004-03-05 | | Release date: | 2004-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery and evaluation of potent P1 aryl heterocycle-based thrombin inhibitors
J.Med.Chem., 47, 2004
|
|
3HZV
 
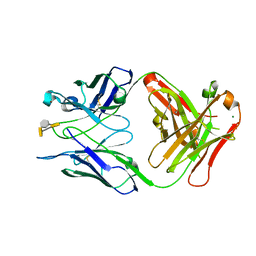 | | Crystal structure of S73-2 antibody in complex with antigen Kdo(2.8)Kdo(2.4)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, MAGNESIUM ION, S73-2 Fab (IgG1k) heavy chain, ... | | Authors: | Brooks, C.L, Muller-Loennies, S, Borisova, S.N, Brade, L, Kosma, P, Hirama, T, MacKenzie, C.R, Brade, H, Evans, S.V. | | Deposit date: | 2009-06-24 | | Release date: | 2010-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibodies raised against chlamydial lipopolysaccharide antigens reveal convergence in germline gene usage and differential epitope recognition
Biochemistry, 49, 2010
|
|
1KQR
 
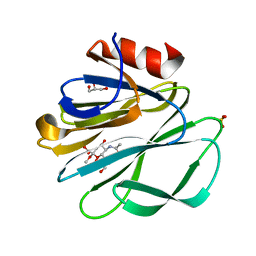 | | Crystal Structure of the Rhesus Rotavirus VP4 Sialic Acid Binding Domain in Complex with 2-O-methyl-alpha-D-N-acetyl neuraminic acid | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Dormitzer, P.R, Sun, Z.-Y.J, Wagner, G, Harrison, S.C. | | Deposit date: | 2002-01-07 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Rhesus Rotavirus VP4 Sialic Acid Binding Domain has a Galectin Fold with a
Novel Carbohydrate Binding Site
Embo J., 21, 2002
|
|
3DW5
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23S rRNA, U2656-OCH3 modified | | Descriptor: | Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
