3V1G
 
 | | Forestalling insulin fibrillation by insertion of a chiral clamp mechanism-based application of protein engineering to global health | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Wan, Z.L, Hua, Q.X, Wickramasinghe, N.P, Huang, K, Petkova, A.T, Hu, S.Q, Phillips, N.B, Yeh, I.J, Whittake, J, Ismail-Beigi, F, Katsoyyannis, P.G, Tycko, R, Weiss, M.A. | | Deposit date: | 2011-12-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Forestalling insulin fibrillation by insertion of a
chiral clamp mechanism-based application of protein engineering to global health
To be Published
|
|
2Q7C
 
 | | Crystal structure of IQN17 | | Descriptor: | CHLORIDE ION, fusion protein between yeast variant GCN4 and HIVgp41 | | Authors: | Malashkevich, V.N, Eckert, D.M, Hong, L.H, Kim, P.S. | | Deposit date: | 2007-06-06 | | Release date: | 2007-06-19 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibiting HIV Entry: Discovery of D-Peptide Inhibitors that Target the Gp41 Coiled-Coil Pocket
Cell(Cambridge,Mass.), 99, 1999
|
|
3QRI
 
 | | The crystal structure of human abl1 kinase domain in complex with DCC-2036 | | Descriptor: | 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, SODIUM ION, Tyrosine-protein kinase ABL1 | | Authors: | Chan, W.W, Wise, S.C, Kaufman, M.D, Ahn, Y.M, Ensinger, C.L, Haack, T, Hood, M.M, Jones, J, Lord, J.W, Lu, W.P, Miller, D, Patt, W.C, Smith, B.D, Petillo, P.A, Rutkoski, T.J, Telikepalli, H, Vogeti, L, Yao, T, Chun, L, Clark, R, Evangelista, P, Gavrilescu, L.C, Lazarides, K, Zaleskas, V.M, Stewart, L.J, Van Etten, R.A, Flynn, D.L. | | Deposit date: | 2011-02-18 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Control Inhibition of the BCR-ABL1 Tyrosine Kinase, Including the Gatekeeper T315I Mutant, by the Switch-Control Inhibitor DCC-2036.
Cancer Cell, 19, 2011
|
|
5BMW
 
 | | Crystal structure of T75V mutant of Triosephosphate isomerase from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Bandyopadhyay, D, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2015-05-23 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Probing the role of highly conserved residues in triosephosphate isomerase - analysis of site specific mutants at positions 64 and 75 in the Plasmodial enzyme
Febs J., 282, 2015
|
|
3MLR
 
 | | Crystal structure of anti-HIV-1 V3 Fab 2557 in complex with a NY5 V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1UX2
 
 | | X-ray structure of acetylcholine binding protein (AChBP) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYLCHOLINE BINDING PROTEIN, ... | | Authors: | Celie, P.H.N, Van Rossum-fikkert, S.E, Van Dijk, W.J, Brejc, K, Smit, A.B, Sixma, T.K. | | Deposit date: | 2004-02-18 | | Release date: | 2004-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nicotine and Carbamylcholine Binding to Nicotinic Acetylcholine Receptors as Studied in Achbp Crystal Structures
Neuron, 41, 2004
|
|
1L3B
 
 | | MT0146, THE PRECORRIN-6Y METHYLTRANSFERASE (CBIT) HOMOLOG FROM M. THERMOAUTOTROPHICUM, C2 SPACEGROUP W/ LONG CELL | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, deTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-26 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of Mt0146/CbiT Suggests that the Putative Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
7VBH
 
 | | Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.H, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-31 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
3HRM
 
 | | Crystal structure of Staphylococcus aureus protein SarZ in sulfenic acid form | | Descriptor: | HTH-type transcriptional regulator sarZ | | Authors: | Poor, C.B, Duguid, E, Rice, P.A, He, C. | | Deposit date: | 2009-06-09 | | Release date: | 2009-07-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus.
J.Biol.Chem., 284, 2009
|
|
2XFJ
 
 | | Human BACE-1 in complex with N-((1S,2R)-3-(((1S)-2-(cyclohexylamino)- 1-methyl-2-oxoethyl)amino)-2-hydroxy-1-(phenylmethyl)propyl)-3-(ethylamino)-5-(2-oxo-1-pyrrolidinyl)benzamide | | Descriptor: | BETA-SECRETASE 1, N-[(1S,2R)-1-benzyl-3-{[(1S)-2-(cyclohexylamino)-1-methyl-2-oxoethyl]amino}-2-hydroxypropyl]-3-(ethylamino)-5-(2-oxopyrrolidin-1-yl)benzamide | | Authors: | Clarke, B, Cutler, L, Demont, E, Dingwall, C, Dunsdon, R, Hawkins, J, Howes, C, Hussain, I, Maile, G, Matico, R, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2010-05-24 | | Release date: | 2010-07-07 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bace-1 Inhibitors Using Novel Edge-to-Face Interaction with Arg-296
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XRF
 
 | | Crystal structure of human uridine phosphorylase 2 | | Descriptor: | GLYCEROL, MAGNESIUM ION, URACIL, ... | | Authors: | Welin, M, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Kouznetsova, E, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Der Berg, S, Wahlberg, E, Weigelt, J, Nordlund, P. | | Deposit date: | 2010-09-14 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Human Uridine Phosphorylase 2
To be Published
|
|
2XCR
 
 | | The 3.5A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase complexed with GSK299423 and DNA | | Descriptor: | 5'-D(*5UA*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP *GP*CP*TP)-3', 5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP *GP*CP*TP)-3', 6-METHOXY-4-(2-{4-[([1,3]OXATHIOLO[5,4-C]PYRIDIN-6-YLMETHYL)AMINO]PIPERIDIN-1-YL}ETHYL)QUINOLINE-3-CARBONITRILE, ... | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-25 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
1BA3
 
 | | FIREFLY LUCIFERASE IN COMPLEX WITH BROMOFORM | | Descriptor: | LUCIFERASE, TRIBROMOMETHANE | | Authors: | Franks, N.P, Jenkins, A, Conti, E, Lieb, W.R, Brick, P. | | Deposit date: | 1998-04-21 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the inhibition of firefly luciferase by a general anesthetic.
Biophys.J., 75, 1998
|
|
2XUF
 
 | | CRYSTAL STRUCTURE OF MACHE-Y337A-TZ2PA6 ANTI COMPLEX (1 MTH) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,8-DIAMINO-6-PHENYL-5-[6-[1-[2-[(1,2,3,4-TETRAHYDRO-9-ACRIDINYL)AMINO]ETHYL]-1H-1,2,3-TRIAZOL-4-YL]HEXYL]-PHENANTHRIDINIUM, ACETYLCHOLINESTERASE, ... | | Authors: | Bourne, Y, Radic, Z, Taylor, P, Marchot, P. | | Deposit date: | 2010-10-19 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Conformational Remodeling of Femtomolar Inhibitor-Acetylcholinesterase Complexes in the Crystalline State
J.Am.Chem.Soc., 132, 2010
|
|
1ZVX
 
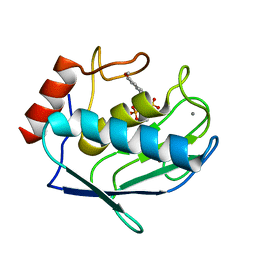 | | Crystal structure of the complex between MMP-8 and a phosphonate inhibitor (R-enantiomer) | | Descriptor: | (1R)-1-{[(4'-METHOXY-1,1'-BIPHENYL-4-YL)SULFONYL]AMINO}-2-METHYLPROPYLPHOSPHONIC ACID, CALCIUM ION, Neutrophil collagenase, ... | | Authors: | Pochetti, G, Gavuzzo, E, Campestre, C, Agamennone, M, Tortorella, P, Consalvi, V, Gallina, C, Hiller, O, Tschesche, H, Tucker, P.A, Mazza, F. | | Deposit date: | 2005-06-03 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural insight into the stereoselective inhibition of MMP-8 by enantiomeric sulfonamide phosphonates.
J.Med.Chem., 49, 2006
|
|
2XYC
 
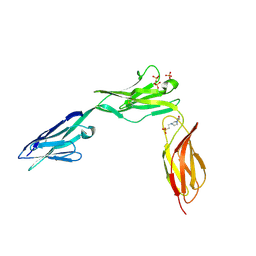 | | CRYSTAL STRUCTURE OF NCAM2 IGIV-FN3I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NEURAL CELL ADHESION MOLECULE 2, ... | | Authors: | Kulahin, N, Rasmussen, K.K, Kristensen, O, Berezin, V, Bock, E, Walmod, P.S, Gajhede, M. | | Deposit date: | 2010-11-17 | | Release date: | 2011-02-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Structure, 19, 2011
|
|
2PZX
 
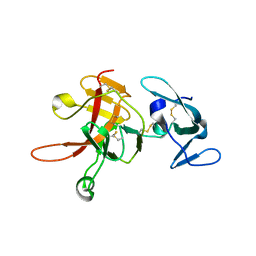 | | Structure of the methuselah ectodomain with peptide inhibitor | | Descriptor: | G-protein coupled receptor Mth | | Authors: | West Jr, A.P, Ja, W.W, Delker, S.L, Bjorkman, P.J, Benzer, S, Roberts, R.W. | | Deposit date: | 2007-05-18 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Extension of Drosophila melanogaster life span with a GPCR peptide inhibitor.
Nat.Chem.Biol., 3, 2007
|
|
4C0Q
 
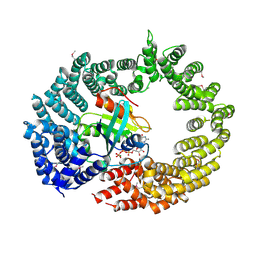 | | Transportin 3 in complex with Ran(Q69L)GTP | | Descriptor: | GTP-BINDING NUCLEAR PROTEIN RAN, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Maertens, G, Hare, S, Cherepanov, P. | | Deposit date: | 2013-08-06 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural Basis for Nuclear Import of Splicing Factors by Human Transportin 3.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6GOO
 
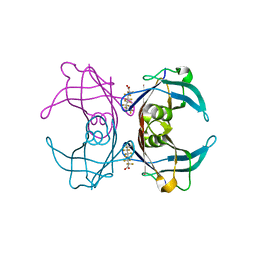 | | Crystal Structure Of Sea Bream Transthyretin in complex with Perfluorooctanoic acid (PFOA). Crystallized in AmSO4 | | Descriptor: | Transthyretin, pentadecafluorooctanoic acid | | Authors: | Grundstrom, C, Zhang, J, Olofsson, A, Andersson, P.L, Sauer-Eriksson, A.E. | | Deposit date: | 2018-06-01 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interspecies Variation between Fish and Human Transthyretins in Their Binding of Thyroid-Disrupting Chemicals.
Environ. Sci. Technol., 52, 2018
|
|
1ZSV
 
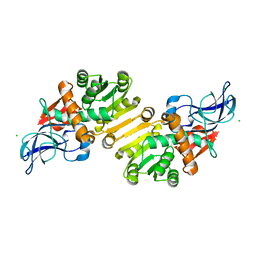 | | Crystal structure of human NADP-dependent leukotriene B4 12-hydroxydehydrogenase | | Descriptor: | CHLORIDE ION, NADP-dependent leukotriene B4 12-hydroxydehydrogenase | | Authors: | Turnbull, A.P, Johansson, C, Savitsky, P, Guo, K, Edwards, A, Arrowsmith, C, Sundstrom, M, von Delft, F, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-05-25 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human NADP-dependent leukotriene B4 12-hydroxydehydrogenase
To be Published
|
|
1LOV
 
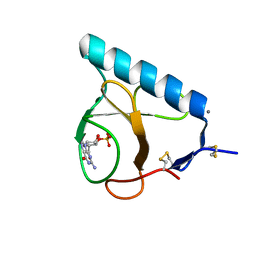 | | X-ray structure of the E58A mutant of Ribonuclease T1 complexed with 3'-guanosine monophosphate | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, Guanyl-specific ribonuclease T1 | | Authors: | Mignon, P, Steyaert, J, Loris, R, Geerlings, P, Loverix, S. | | Deposit date: | 2002-05-07 | | Release date: | 2002-08-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A nucleophile activation dyad in ribonucleases. A combined X-ray crystallographic/ab initio quantum chemical study
J.Biol.Chem., 277, 2002
|
|
3M3L
 
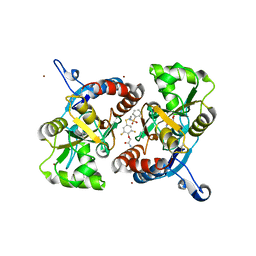 | | PEPA bound to the ligand binding domain of GluA2 (flop form) | | Descriptor: | 2-[2,6-difluoro-4-({2-[(phenylsulfonyl)amino]ethyl}sulfanyl)phenoxy]acetamide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ptak, C.P, Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular mechanism of flop selectivity and subsite recognition for an AMPA receptor allosteric modulator: structures of GluA2 and GluA3 in complexes with PEPA.
Biochemistry, 49, 2010
|
|
3HPG
 
 | |
2XL4
 
 | | LntA, a virulence factor from Listeria monocytogenes | | Descriptor: | GLYCEROL, Listeria nuclear targeted protein A | | Authors: | Lebreton, A, Job, V, Tham, T.N, Camejo, A, Mattei, P.J, Regnault, B, Cabanes, D, Dessen, A, Cossart, P, Bierne, H. | | Deposit date: | 2010-07-19 | | Release date: | 2011-02-02 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response.
Science, 331, 2011
|
|
3HQ2
 
 | | BsuCP Crystal Structure | | Descriptor: | Bacillus subtilis M32 carboxypeptidase, CHLORIDE ION, FLUORIDE ION, ... | | Authors: | Lee, M.M, Isaza, C.E, White, J.D, Chen, R.P.-Y, Liang, G.F.-C, He, H.T.-F, Chan, S.I, Chan, M.K. | | Deposit date: | 2009-06-05 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insight into the substrate length restriction of M32 carboxypeptidases: Characterization of two distinct subfamilies.
Proteins, 77, 2009
|
|
