8OLA
 
 | | Carbonic Anhydrase IX like mutant in Complex with Steriod_Sulphamoyl inhibitor VK4 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{R},5~{R},8~{R},9~{S},10~{S},13~{S},14~{S},17~{S})-17-ethanoyl-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
8OMH
 
 | | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl (BS1982) | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(8~{S},9~{S},13~{S},14~{S})-13-methyl-6,7,8,9,11,12,14,15,16,17-decahydrocyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-31 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
1GEC
 
 | | GLYCYL ENDOPEPTIDASE-COMPLEX WITH BENZYLOXYCARBONYL-LEUCINE-VALINE-GLYCINE-METHYLENE COVALENTLY BOUND TO CYSTEINE 25 | | Descriptor: | BENZYLOXYCARBONYL-LEUCINE-VALINE-GLYCINE-METHYLENE INHIBITOR, GLYCYL ENDOPEPTIDASE | | Authors: | Ohara, B.P, Hemmings, A.M, Buttle, D.J, Pearl, L.H. | | Deposit date: | 1995-05-25 | | Release date: | 1995-12-07 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of glycyl endopeptidase from Carica papaya: a cysteine endopeptidase of unusual substrate specificity.
Biochemistry, 34, 1995
|
|
8OKO
 
 | | Carbonic Anhydrase IX like mutant in Complex with Steriod_Sulphamoyl inhibitor AKI_2 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{S},8~{S},9~{S},10~{R},13~{S},14~{S})-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
4E7Y
 
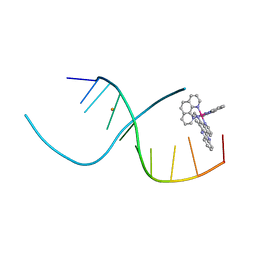 | | Lambda-[Ru(phen)2(dppz)]2+ Bound to CCGGATCCGG | | Descriptor: | 5'-D(*CP*CP*GP*GP*AP*TP*CP*CP*GP*G)-3', BARIUM ION, Lambda-Ru(phen)2(dppz) complex | | Authors: | Hall, J.P, Cardin, C.J. | | Deposit date: | 2012-03-19 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of lambda-[Ru(phen)2(dppz)]2+ with oligonucleotides containing TA/TA and AT/AT steps show two intercalation modes.
Nat Chem, 4, 2012
|
|
8OKT
 
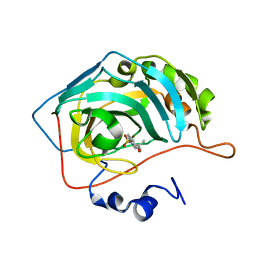 | | Carbonic Anhydrase IX like mutant in Complex with Steriod_Sulphamoyl inhibitor VK42 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(8~{R},9~{S},13~{S},14~{S})-13-methyl-17-oxidanylidene-7,8,9,11,12,14,15,16-octahydro-6~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
7X2Y
 
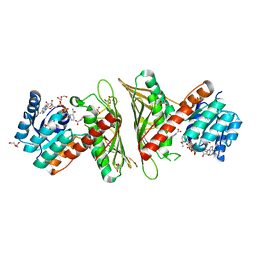 | | Crystal Structure of cis-4,5-dihydrodiol phthalate dehydrogenase in complex with NAD+ and 3-Hydroxybenzoate | | Descriptor: | 3-HYDROXYBENZOIC ACID, 4,5-dihydroxyphthalate dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2022-02-26 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conformational flexibility enables catalysis of phthalate cis-4,5-dihydrodiol dehydrogenase.
Arch.Biochem.Biophys., 727, 2022
|
|
8OLM
 
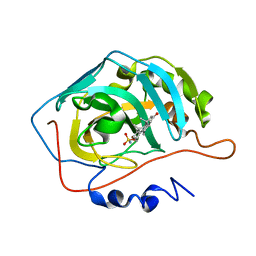 | | Carbonic Anhydrase 2 in Complex with Steriod_Sulphamoyl AKI1 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{S},5~{S},8~{S},13~{R},14~{R})-5,14-dimethyl-16-oxidanylidene-1,2,3,4,6,7,8,11,12,13,15,17-dodecahydrocyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Carbonic Anhydrase in Complex with Steriod_Sulphamoyl
To Be Published
|
|
1YG3
 
 | |
8OMB
 
 | | Carbonic Anhydrase 2 in Complex with Steriod_Sulphamoyl VK42 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(8~{R},9~{S},13~{S},14~{S})-13-methyl-17-oxidanylidene-7,8,9,11,12,14,15,16-octahydro-6~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-31 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Carbonic Anhydrase in Complex with Steriod_Sulphamoyl
To Be Published
|
|
4E7K
 
 | | PFV integrase Target Capture Complex (TCC-Mn), freeze-trapped prior to strand transfer, at 3.0 A resolution | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*CP*CP*CP*GP*AP*GP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*CP*TP*CP*GP*GP*G)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Maertens, G.N, Cherepanov, P. | | Deposit date: | 2012-03-17 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | 3'-Processing and strand transfer catalysed by retroviral integrase in crystallo.
Embo J., 31, 2012
|
|
8OMN
 
 | | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl VK4 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{R},5~{R},8~{R},9~{S},10~{S},13~{S},14~{S},17~{S})-17-ethanoyl-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-31 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
5ORO
 
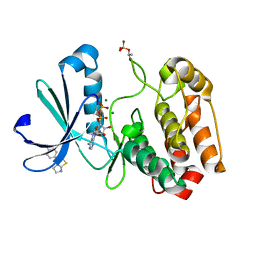 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | 3-(4-chlorophenyl)-5,6-dihydroimidazo[2,1-b][1,3]thiazole, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | McIntyre, P.J, Collins, P.M, von Delft, F, Bayliss, R. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-01 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
5ORR
 
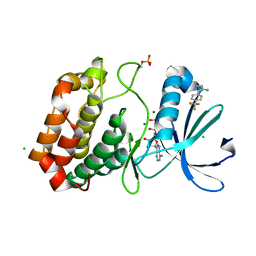 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | 4-[4-(trifluoromethyl)phenyl]-1,2,3-thiadiazol-5-amine, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | McIntyre, P.J, Collins, P.M, von Delft, F, Bayliss, R. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-01 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
1FY4
 
 | | FUSARIUM OXYSPORUM TRYPSIN AT ATOMIC RESOLUTION | | Descriptor: | GLY-ALA-ARG, GLYCEROL, SULFATE ION, ... | | Authors: | Rypniewski, W.R, Oestergaard, P, Noerregaard-Madsen, M, Dauter, M, Wilson, K.S. | | Deposit date: | 2000-09-28 | | Release date: | 2001-02-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.81 Å) | | Cite: | Fusarium oxysporum trypsin at atomic resolution at 100 and 283 K: a study of ligand binding.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5EIR
 
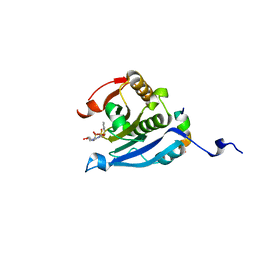 | | Co-crystal structure of eIF4E with nucleotide mimetic inhibitor. | | Descriptor: | Eukaryotic translation initiation factor 4 gamma 1, Eukaryotic translation initiation factor 4E, SULFATE ION, ... | | Authors: | Nowicki, M.W, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2015-10-30 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design of nucleotide-mimetic and non-nucleotide inhibitors of the translation initiation factor eIF4E: Synthesis, structural and functional characterisation.
Eur.J.Med.Chem., 124, 2016
|
|
1G2O
 
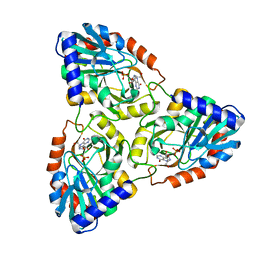 | | CRYSTAL STRUCTURE OF PURINE NUCLEOSIDE PHOSPHORYLASE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH A TRANSITION-STATE INHIBITOR | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PHOSPHATE ION, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Shi, W, Basso, L.A, Tyler, P.C, Furneaux, R.H, Blanchard, J.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2000-10-20 | | Release date: | 2001-08-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of purine nucleoside phosphorylase from Mycobacterium tuberculosis in complexes with immucillin-H and its pieces.
Biochemistry, 40, 2001
|
|
6JCZ
 
 | | Cryo-EM Structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) in complex with Mg2+, NADPH, and CPD at pH7.5 | | Descriptor: | MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative ketol-acid reductoisomerase 2, ... | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
5U75
 
 | | The structure of Staphylococcal Enterotoxin-like X (SElX), a Unique Superantigen | | Descriptor: | Enterotoxin-like toxin X, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Ting, Y.T, Young, P.G, Langley, R.J, Baker, H, Fraser, J.D. | | Deposit date: | 2016-12-11 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Staphylococcal enterotoxin-like X (SElX) is a unique superantigen with functional features of two major families of staphylococcal virulence factors.
PLoS Pathog., 13, 2017
|
|
1GAM
 
 | | GAMMA B CRYSTALLIN TRUNCATED C-TERMINAL DOMAIN | | Descriptor: | GAMMA B CRYSTALLIN | | Authors: | Norledge, B.V, Mayr, E.-M, Glockshuber, R, Bateman, O.A, Slingsby, C, Jaenicke, R, Driessen, H.P.C. | | Deposit date: | 1996-02-02 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The X-ray structures of two mutant crystallin domains shed light on the evolution of multi-domain proteins.
Nat.Struct.Biol., 3, 1996
|
|
8A9L
 
 | | Cryo-EM structure of alpha-synuclein filaments from Parkinson's disease and dementia with Lewy bodies | | Descriptor: | Alpha-synuclein, Unknown fragment | | Authors: | Yang, Y, Shi, Y, Schweighauser, M, Zhang, X.J, Kotecha, A, Murzin, A.G, Garringer, H.J, Cullinane, P, Saito, Y, Foroud, T, Warner, T.T, Hasegawa, K, Vidal, R, Murayama, S, Revesz, T, Ghetti, B, Hasegawa, M, Lashley, T, Scheres, H.W.S, Goedert, M. | | Deposit date: | 2022-06-28 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structures of alpha-synuclein filaments from human brains with Lewy pathology.
Nature, 610, 2022
|
|
2CCY
 
 | | STRUCTURE OF FERRICYTOCHROME C(PRIME) FROM RHODOSPIRILLUM MOLISCHIANUM AT 1.67 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Finzel, B.C, Weber, P.C, Hardman, K.D, Salemme, F.R. | | Deposit date: | 1985-08-27 | | Release date: | 1986-01-21 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure of ferricytochrome c' from Rhodospirillum molischianum at 1.67 A resolution.
J.Mol.Biol., 186, 1985
|
|
4WGX
 
 | | Crystal Structure of Molinate Hydrolase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COBALT (II) ION, Molinate hydrolase | | Authors: | Sugrue, E, Carr, P.D, Fraser, N.J, Hopkins, D.H, Jackson, C.J. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Evolutionary Expansion of the Amidohydrolase Superfamily in Bacteria in Response to the Synthetic Compounds Molinate and Diuron.
Appl.Environ.Microbiol., 81, 2015
|
|
5NPZ
 
 | | Porcine (Sus scrofa) Major Histocompatibility Complex, class I, presenting EFEDLTFLA | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rizkallah, P.J, Tungatt, K, Sewell, A.K. | | Deposit date: | 2017-04-19 | | Release date: | 2018-05-02 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Induction of influenza-specific local CD8 T-cells in the respiratory tract after aerosol delivery of vaccine antigen or virus in the Babraham inbred pig.
Plos Pathog., 14, 2018
|
|
6NMZ
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with an intentionally produced fragment degradation product B9D | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, SULFATE ION, [(1R,2S)-2-(2-hydroxybenzene-1-carbonyl)cyclopentyl]acetic acid, ... | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-01-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with an intentionally produced fragment degradation product B9D
To Be Published
|
|
