4TKI
 
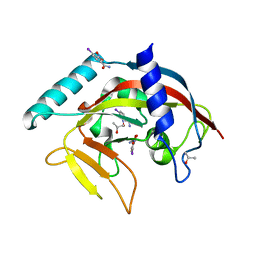 | | Crystal Structure of human Tankyrase 2 in complex with BSI-201. | | Descriptor: | 4-iodo-3-nitrobenzamide, ISOPROPYL ALCOHOL, Tankyrase-2, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8HRG
 
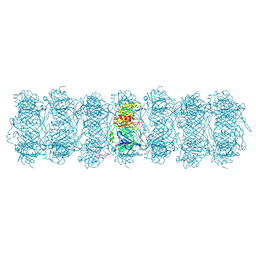 | | Tail tube of DT57C bacteriophage in the full state | | Descriptor: | Tail tube protein | | Authors: | Ayala, R, Moiseenko, A.V, Chen, T.H, Kulikov, E.E, Golomidova, A.K, Orekhov, P.S, Street, M.A, Sokolova, O.S, Letarov, A.V, Wolf, M. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers.
Nat Commun, 14, 2023
|
|
6U7W
 
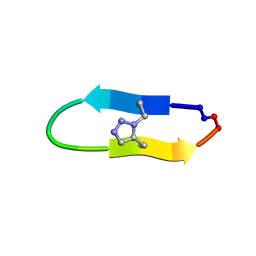 | | NMR solution structure of a triazole bridged KLK7 inhibitor | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-LYS-ALA-LEU-PHE-SER-ASN-PRO-PRO-ILE-ALA-PHE-PRO-ASN | | Authors: | White, A.M, Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2020-07-15 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8HQK
 
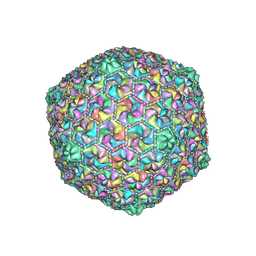 | | Capsid of DT57C bacteriophage in the empty state | | Descriptor: | Major head protein | | Authors: | Ayala, R, Moiseenko, A.V, Kulikov, E.E, Golomidova, A.K, Orekhov, P.S, Street, M.A, Sokolova, O.S, Letarov, A.V, Wolf, M. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers.
Nat Commun, 14, 2023
|
|
3HKV
 
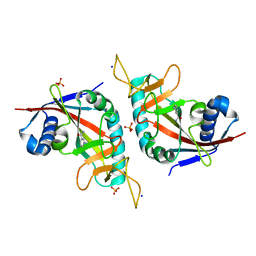 | | Human poly(ADP-ribose) polymerase 10, catalytic fragment in complex with an inhibitor 3-aminobenzamide | | Descriptor: | 3-aminobenzamide, PHOSPHATE ION, Poly [ADP-ribose] polymerase 10, ... | | Authors: | Karlberg, T, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Kotzsch, A, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Sagemark, J, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-26 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Poly(Adp-Ribose) Polymerase 10, Catalytic Fragment in Complex with an Inhibitor 3-Aminobenzamide
To be Published
|
|
1CE7
 
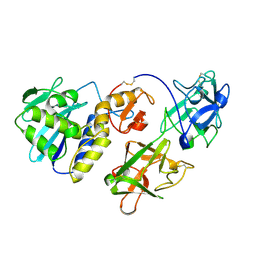 | | MISTLETOE LECTIN I FROM VISCUM ALBUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (RIBOSOME-INACTIVATING PROTEIN TYPE II) | | Authors: | Krauspenhaar, R, Eschenburg, S, Perbandt, M, Kornilov, V, Konareva, N, Mikailova, I, Stoeva, S, Wacker, R, Maier, T, Singh, T.P, Mikhailov, A, Voelter, W, Betzel, C. | | Deposit date: | 1999-03-18 | | Release date: | 2000-03-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of mistletoe lectin I from Viscum album.
Biochem.Biophys.Res.Commun., 257, 1999
|
|
5GVM
 
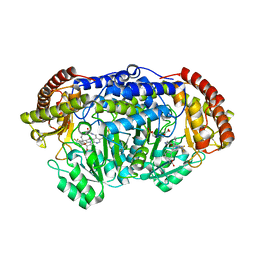 | | Plasmodium vivax SHMT bound with PLP-glycine and GS557 | | Descriptor: | 2-[3-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-(trifluoromethyl)phenyl]phenyl]ethanoic acid, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G. | | Deposit date: | 2016-09-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Antimalarial Inhibitors Targeting Serine Hydroxymethyltransferase (SHMT) with in Vivo Efficacy and Analysis of their Binding Mode Based on X-ray Cocrystal Structures
J. Med. Chem., 60, 2017
|
|
1D6F
 
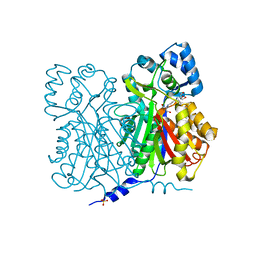 | | CHALCONE SYNTHASE C164A MUTANT | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHALCONE SYNTHASE, SULFATE ION | | Authors: | Jez, J.M, Ferrer, J.L, Bowman, M.E, Dixon, R.A, Noel, J.P. | | Deposit date: | 1999-10-13 | | Release date: | 2000-02-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase.
Biochemistry, 39, 2000
|
|
2I0Q
 
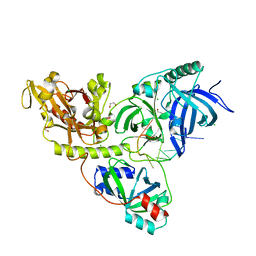 | |
8HO3
 
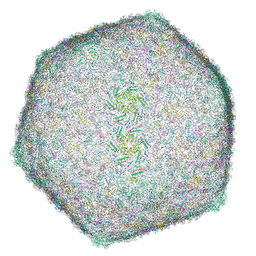 | | Capsid of DT57C bacteriophage in the full state | | Descriptor: | Major head protein | | Authors: | Ayala, R, Moiseenko, A.V, Chen, T.H, Kulikov, E.E, Golomidova, A.K, Orekhov, P.S, Street, M.A, Sokolova, O.S, Letarov, A.V, Wolf, M. | | Deposit date: | 2022-12-09 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers.
Nat Commun, 14, 2023
|
|
5EIH
 
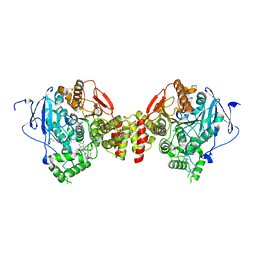 | | mAChE-TZ2/PA5 complex | | Descriptor: | 5-hept-6-ynyl-6-phenyl-phenanthridin-5-ium-3,8-diamine, ACETATE ION, Acetylcholinesterase, ... | | Authors: | Bourne, Y, Marchot, P. | | Deposit date: | 2015-10-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Steric and Dynamic Parameters Influencing In Situ Cycloadditions to Form Triazole Inhibitors with Crystalline Acetylcholinesterase.
J.Am.Chem.Soc., 138, 2016
|
|
6FUN
 
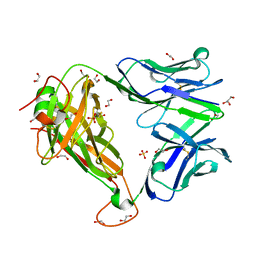 | |
5JGU
 
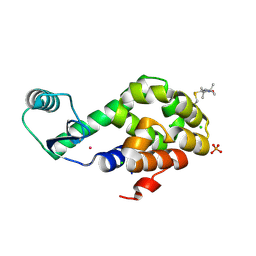 | | Spin-Labeled T4 Lysozyme Construct R119V1 | | Descriptor: | CHLORIDE ION, Endolysin, PHOSPHATE ION, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.468 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
1H03
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-06-11 | | Release date: | 2003-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
1UJL
 
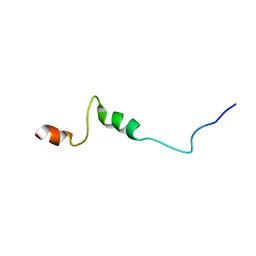 | | Solution Structure of the HERG K+ channel S5-P extracellular linker | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Torres, A.M, Bansal, P.S, Sunde, M, Clarke, C.E, Bursill, J.A, Smith, D.J, Bauskin, A, Breit, S.N, Campbell, T.J, Alewood, P.F, Kuchel, P.W, Vandenberg, J.I. | | Deposit date: | 2003-08-05 | | Release date: | 2003-11-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the HERG K+ channel S5P extracellular linker: role of an amphipathic alpha-helix
in C-type inactivation.
J.Biol.Chem., 278, 2003
|
|
8J5J
 
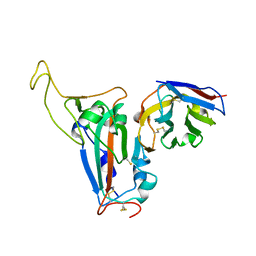 | | The crystal structure of bat coronavirus RsYN04 RBD bound to the antibody S43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, nano antibody S43 | | Authors: | Zhao, R.C, Niu, S, Han, P, Qi, J.X, Gao, G.F, Wang, Q.H. | | Deposit date: | 2023-04-23 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cross-species recognition of bat coronavirus RsYN04 and cross-reaction of SARS-CoV-2 antibodies against the virus.
Zool.Res., 44, 2023
|
|
4GV1
 
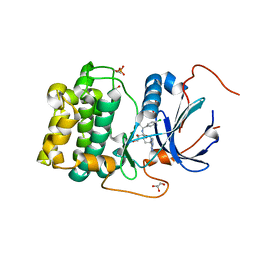 | | PKB alpha in complex with AZD5363 | | Descriptor: | 4-amino-N-[(1S)-1-(4-chlorophenyl)-3-hydroxypropyl]-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidine-4-carboxamide, GLYCEROL, RAC-alpha serine/threonine-protein kinase | | Authors: | Addie, M, Ballard, P, Bird, G, Buttar, D, Currie, G, Davies, B, Debreczeni, J, Dry, H, Dudley, P, Greenwood, R, Hatter, G, Jestel, A, Johnson, P.D, Kettle, J.G, Lane, C, Lamont, G, Leach, A, Luke, R.W.A, Ogilvie, D, Page, K, Pass, M, Steinbacher, S, Steuber, H, Pearson, S, Ruston, L. | | Deposit date: | 2012-08-30 | | Release date: | 2013-02-27 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of 4-Amino-N-[(1S)-1-(4-chlorophenyl)-3-hydroxypropyl]-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidine-4-carboxamide (AZD5363), an Orally Bioavailable, Potent Inhibitor of Akt Kinases.
J.Med.Chem., 56, 2013
|
|
5D9F
 
 | |
5JJZ
 
 | | Chromo domain of human Chromodomain Protein, Y-Like 2 | | Descriptor: | Chromodomain Y-like protein 2, LYS-LYS-LYS-ALA-ARG-MLY-SER-ALA-GLY-ALA-ALA-LYS-TYR | | Authors: | DONG, A, DOMBROVSKI, L, LOPPNAU, P, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, MIN, J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-04-25 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of CDYL2 domain of human CDYL2 protein
to be published
|
|
1L3M
 
 | | The Solution Structure of [d(CGC)r(amamam)d(TTTGCG)]2 | | Descriptor: | 5'-D(*CP*GP*C)-R(P*(A39)P*(A39)P*(A39))-D(P*TP*TP*TP*GP*CP*G)-3' | | Authors: | Tsao, Y.P, Wang, L.Y, Hsu, S.T, Jain, M.L, Chou, S.H, Huang, W.C, Cheng, J.W. | | Deposit date: | 2002-02-28 | | Release date: | 2002-04-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of [d(CGC)r(amamam)d(TTTGCG)]2.
J.Biomol.NMR, 21, 2001
|
|
2O4M
 
 | | Structure of Phosphotriesterase mutant I106G/F132G/H257Y | | Descriptor: | ACETIC ACID, CACODYLATE ION, GLYCEROL, ... | | Authors: | Kim, J, Ramagopal, U.A, Tsai, P, Raushel, F.M, Almo, S.C. | | Deposit date: | 2006-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of Phosphotriesterase mutant I106G/F132G/H257Y
To be Published
|
|
5JJM
 
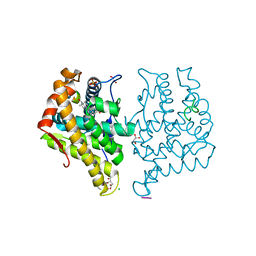 | |
8HLT
 
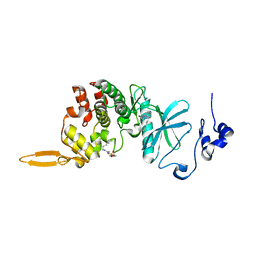 | | The co-crystal structure of DYRK2 with YK-2-99B | | Descriptor: | (6-{[(4P)-4-(1,3-benzothiazol-5-yl)-5-fluoropyrimidin-2-yl]amino}pyridin-3-yl)(piperazin-1-yl)methanone, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Shen, H.T, Xiao, Y.B, Yuan, K, Yang, P, Li, Q.N. | | Deposit date: | 2022-12-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Potent DYRK2 Inhibitors with High Selectivity, Great Solubility, and Excellent Safety Properties for the Treatment of Prostate Cancer.
J.Med.Chem., 66, 2023
|
|
4TSH
 
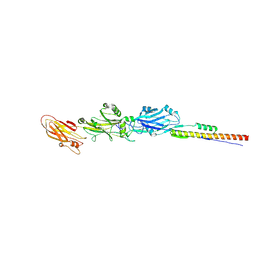 | | A Novel Protein Fold Forms an Intramolecular Lock to Stabilize the Tertiary Structure of Streptococcus mutans Adhesin P1 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Surface protein adhesin | | Authors: | Heim, K.P, Kailasan, S, McKenna, R, Brady, L.J. | | Deposit date: | 2014-06-18 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An intramolecular lock facilitates folding and stabilizes the tertiary structure of Streptococcus mutans adhesin P1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6T58
 
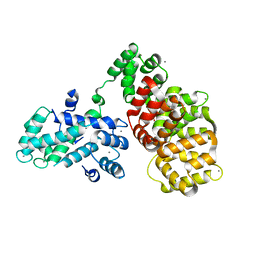 | | Structure determination of the transactivation domain of p53 in complex with S100A4 using annexin A2 as a crystallization chaperone | | Descriptor: | CALCIUM ION, Cellular tumor antigen p53,Protein S100-A4,Protein S100-A4,Annexin A2, GLYCEROL | | Authors: | Ecsedi, P, Gogl, G, Nyitray, L. | | Deposit date: | 2019-10-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure Determination of the Transactivation Domain of p53 in Complex with S100A4 Using Annexin A2 as a Crystallization Chaperone.
Structure, 28, 2020
|
|
