5REB
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z2856434899 | | Descriptor: | 1-[(thiophen-3-yl)methyl]piperidin-4-ol, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
7C75
 
 | | Crystal structure of yak lactoperoxidase with partially coordinated Na ion in the distal heme cavity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Lactoperoxidase, ... | | Authors: | Singh, P.K, Viswanathan, V, Rani, C, Ahmad, N, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Potassium-induced partial inhibition of lactoperoxidase: structure of the complex of lactoperoxidase with potassium ion at 2.20 angstrom resolution.
J.Biol.Inorg.Chem., 26, 2021
|
|
5RGO
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102248 (Mpro-x0736) | | Descriptor: | 1-[4-(furan-2-carbonyl)piperazin-1-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-15 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
7TXD
 
 | | Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Broadly neutralizing darpin bnd.9, ... | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2022-02-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Trapping the HIV-1 V3 loop in a helical conformation enables broad neutralization.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5RF2
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1741969146 | | Descriptor: | 1-azanylpropylideneazanium, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
3I3N
 
 | | Crystal structure of the BTB-BACK domains of human KLHL11 | | Descriptor: | CHLORIDE ION, Kelch-like protein 11, THIOCYANATE ION | | Authors: | Murray, J.W, Cooper, C.D.O, Krojer, T, Mahajan, P, Salah, E, Keates, T, Savitsky, P, Pike, A.C.W, Roos, A, Muniz, J, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the BTB-BACK domains of human KLHL11
To be Published
|
|
6Z9Y
 
 | | Copper transporter OprC | | Descriptor: | COPPER (II) ION, Putative copper transport outer membrane porin OprC | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
5RFG
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102372 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(3S)-1,1-dioxo-2,3-dihydro-1H-1lambda~6~-thiophen-3-yl]-N-phenylacetamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFU
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102121 | | Descriptor: | 1-{4-[(5-chlorothiophen-2-yl)sulfonyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
6UDC
 
 | | Spectroscopic and structural characterization of a genetically encoded direct sensor for protein-ligand interactions | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Mills, J.H, Gleason, P.R, Simmons, C.R, Henderson, J.N, Kartchner, B.K. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Origins of Altered Spectroscopic Properties upon Ligand Binding in Proteins Containing a Fluorescent Noncanonical Amino Acid.
Biochemistry, 60, 2021
|
|
5RHF
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with PG-COV-34 (Mpro-x2754) | | Descriptor: | 1-acetyl-N-methyl-N-phenylpiperidine-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-16 | | Release date: | 2020-06-10 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
6OHK
 
 | |
7Y5V
 
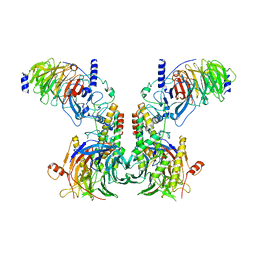 | | Cryo-EM structure of the dimeric human CAF1LC-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, C, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
4WO7
 
 | |
7Y5U
 
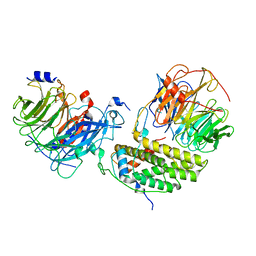 | | Cryo-EM structure of the monomeric human CAF1LC-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
6ZGR
 
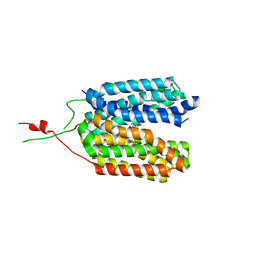 | | Crystal structure of a MFS transporter with bound 1-hydroxynaphthalene-2-carboxylic acid at 2.67 Angstroem resolution | | Descriptor: | 1-hydroxynaphthalene-2-carboxylic acid, L-lactate transporter | | Authors: | Kalbermatter, D, Bosshart, P, Bonetti, S, Fotiadis, D. | | Deposit date: | 2020-06-19 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The making of a potent L-lactate transport inhibitor
Commun Chem, 4, 2021
|
|
6I24
 
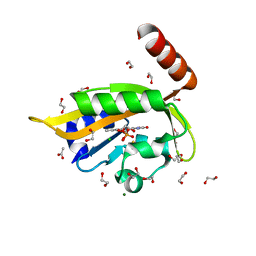 | | Flavin Analogue Sheds Light on Light-Oxygen-Voltage Domain Mechanism | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-5-O-phosphono-D-ribitol, ACETATE ION, ... | | Authors: | Rizkallah, P.J, Kalvaitis, M.E, Allemann, R.K, Mart, R.J, Johnson, L.A. | | Deposit date: | 2018-10-31 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A Noncanonical Chromophore Reveals Structural Rearrangements of the Light-Oxygen-Voltage Domain upon Photoactivation.
Biochemistry, 58, 2019
|
|
7YIX
 
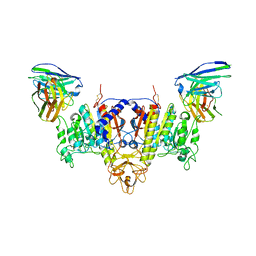 | | The Cryo-EM Structure of Human Tissue Nonspecific Alkaline Phosphatase and Single-Chain Fragment Variable (ScFv) Complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, ... | | Authors: | Yu, Y.T, Yao, D.Q, Zhang, Q, Rao, B, Xia, Y, Lu, Y, Qin, A, Ma, P.X, Cao, Y. | | Deposit date: | 2022-07-18 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | The structural pathology for hypophosphatasia caused by malfunctional tissue non-specific alkaline phosphatase.
Nat Commun, 14, 2023
|
|
6ZGU
 
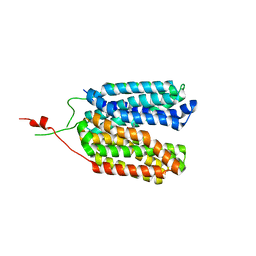 | | Crystal structure of a MFS transporter with bound 3-(2-methylphenyl)propanoic acid at 2.41 Angstroem resolution | | Descriptor: | 3-(2-methylphenyl)propanoic acid, L-lactate transporter | | Authors: | Kalbermatter, D, Bosshart, P, Bonetti, S, Fotiadis, D. | | Deposit date: | 2020-06-19 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The making of a potent L-lactate transport inhibitor
Commun Chem, 4, 2021
|
|
4WQ2
 
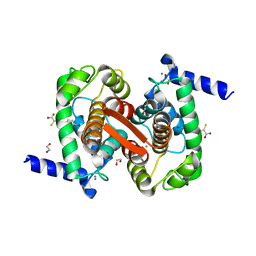 | | Human calpain PEF(S) with (Z)-3-(6-bromondol-3-yl)-2-mercaptoacrylic acid bound | | Descriptor: | (2Z)-3-(6-bromo-1H-indol-3-yl)-2-sulfanylprop-2-enoic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Adams, S.E, Rizkallah, P.J, Miller, D.J, Hallett, M.B, Allemann, R.K. | | Deposit date: | 2014-10-21 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Conformationally restricted calpain inhibitors.
Chem Sci, 6, 2015
|
|
6ZGT
 
 | | Crystal structure of a MFS transporter with bound 2-naphthoic acid at 2.39 Angstroem resolution | | Descriptor: | L-lactate transporter, naphthalene-2-carboxylic acid | | Authors: | Kalbermatter, D, Bosshart, P, Bonetti, S, Fotiadis, D. | | Deposit date: | 2020-06-19 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The making of a potent L-lactate transport inhibitor
Commun Chem, 4, 2021
|
|
6ZGS
 
 | | Crystal structure of a MFS transporter with bound 3-phenylpropanoic acid at 2.39 Angstroem resolution | | Descriptor: | HYDROCINNAMIC ACID, L-lactate transporter | | Authors: | Kalbermatter, D, Bosshart, P, Bonetti, S, Fotiadis, D. | | Deposit date: | 2020-06-19 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | The making of a potent L-lactate transport inhibitor
Commun Chem, 4, 2021
|
|
4ZPV
 
 | | Structure of MERS-Coronavirus Spike Receptor-binding Domain (England1 Strain) in Complex with Vaccine-Elicited Murine Neutralizing Antibody D12 (Crystal Form 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D12 Fab Heavy chain, D12 Fab light chain, ... | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Evaluation of candidate vaccine approaches for MERS-CoV.
Nat Commun, 6, 2015
|
|
5KJW
 
 | | Crystal structure of Coleus blumei HCT in complex with 3-hydroxyacetophenone | | Descriptor: | 1-(3-hydroxyphenyl)ethanone, Hydroxycinnamoyl transferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KT8
 
 | |
