8RKU
 
 | | Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K TnsC domain local-refinement map | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Non-target strand - LE, ... | | Authors: | Tenjo-Castano, F, Mesa, P, Montoya, G. | | Deposit date: | 2023-12-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
8RKT
 
 | | Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K Cas12k domain local-refinement map | | Descriptor: | MAGNESIUM ION, Non-target strand - LE, ShCas12k, ... | | Authors: | Tenjo-Castano, F, Mesa, P, Montoya, G. | | Deposit date: | 2023-12-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
2G3P
 
 | |
1MU8
 
 | | thrombin-hirugen_l-378,650 | | Descriptor: | 2-(6-CHLORO-3-{[2,2-DIFLUORO-2-(2-PYRIDINYL)ETHYL]AMINO}-2-OXO-1(2H)-PYRAZINYL)-N-[(2-FLUORO-3-METHYL-6-PYRIDINYL)METHYL]ACETAMIDE, HIRUDIN IIB, THROMBIN | | Authors: | Burgey, C.S, Robinson, K.A, Lyle, T.A, Sanderson, P.E, Lewis, S.D, Lucas, B.J, Krueger, J.A, Singh, R, Miller-Stein, C, White, R.B, Wong, B, Lyle, E.A, Williams, P.D, Coburn, C.A, Dorsey, B.D, Barrow, J.C, Stranieri, M.T, Holahan, M.A, Sitko, G.R, Cook, J.J, McMasters, D.R, McDonough, C.M, Sanders, W.M, Wallace, A.A, Clayton, F.C, Bohn, D, Leonard, Y.M, Detwiler Jr, T.J, Lynch Jr, J.J, Yan, Y, Chen, Z, Kuo, L, Gardell, S.J, Shafer, J.A, Vacca, J.P.J. | | Deposit date: | 2002-09-23 | | Release date: | 2004-04-06 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metabolism-directed optimization of 3-aminopyrazinone acetamide thrombin inhibitors. Development of an orally bioavailable series containing P1 and P3 pyridines.
J.Med.Chem., 46, 2003
|
|
7RKV
 
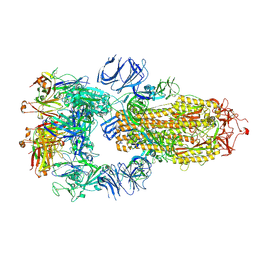 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C118 (State 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C118 Fab Heavy Chain, C118 Fab Light Chain, ... | | Authors: | Barnes, C.O, Jette, C.A, Bjorkman, P.J. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
7RKU
 
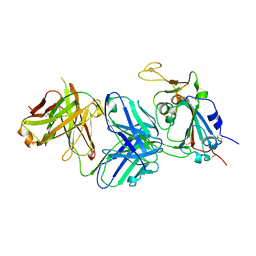 | | Structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment, C022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C022 Antibody Fab Heavy Chain, C022 Antibody Fab Light Chain, ... | | Authors: | Jette, C.A, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
3UIL
 
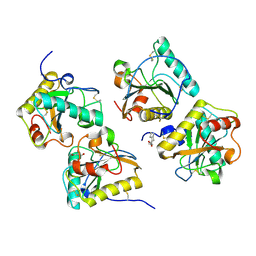 | | Crystal Structure of the complex of PGRP-S with lauric acid at 2.2 A resolution | | Descriptor: | GLYCEROL, LAURIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-05 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
6DMJ
 
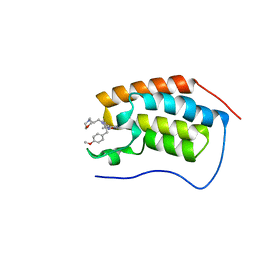 | | A multiconformer ligand model of inhibitor 53W bound to CREB binding protein bromodomain | | Descriptor: | 5-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[2-(4-methoxyphenyl)ethyl]-1-[2-(morpholin-4-yl)ethyl]-1H-benzimidazole, Bromodomain-containing protein 4 | | Authors: | Hudson, B.M, van Zundert, G, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
3V01
 
 | | Discovery of Novel Allosteric MEK Inhibitors Possessing Classical and Non-classical Bidentate Ser212 Interactions. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Heald, R, Jackson, P, Savy, P, Jones, M, Gancia, E, Burton, B, Newman, R, Boggs, J, Chan, E, Chan, J, Choo, E, Merchant, M, Ultsch, M, Wiesmann, C, Belvin, M, Price, S. | | Deposit date: | 2011-12-07 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Discovery of Novel Allosteric Mitogen-Activated Protein Kinase Kinase (MEK) 1,2 Inhibitors Possessing Bidentate Ser212 Interactions.
J.Med.Chem., 55, 2012
|
|
5KCS
 
 | | Cryo-EM structure of the Escherichia coli 70S ribosome in complex with antibiotic Evernimycin, mRNA, TetM and P-site tRNA at 3.9A resolution | | Descriptor: | (2R,3R,4R,6S)-6-{[(2R,3aR,4R,4'R,5'S,6S,6'R,7S,7aR)-6-{[(2S,3R,4R,5S,6R)-2-{[(2R,3S,4S,5S,6S)-6-({(2R,3aS,3a'R,6S,7R,7'R,7aS,7a'S)-7'-[(2,4-dihydroxy-6-methylbenzoyl)oxy]-7-hydroxyoctahydro-4H-2,4'-spirobi[[1,3]dioxolo[4,5-c]pyran]-6-yl}oxy)-4-hydroxy-5-methoxy-2-(methoxymethyl)tetrahydro-2H-pyran-3-yl]oxy}-3-hydroxy-5-methoxy-6-methyltetrahydro-2H-pyran-4-yl]oxy}-4',7-dihydroxy-4,6',7a-trimethyloctahydro-4H-spiro[1,3-dioxolo[4,5-c]pyran-2,2'-pyran]-5'-yl]oxy}-4-{[(2R,4S,5R,6S)-5-methoxy-4,6-dimethyl-4-nitrotetrahydro-2H-pyran-2-yl]oxy}-2-methyltetrahydro-2H-pyran-3-yl 3,5-dichloro-4-hydroxy-2-methoxy-6-methylbenzoate (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Arenz, S, Juette, M.F, Graf, M, Nguyen, F, Huter, P, Polikanov, Y.S, Blanchard, S.C, Wilson, D.N. | | Deposit date: | 2016-06-06 | | Release date: | 2016-08-17 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4F78
 
 | | Crystal Structure of Vancomycin Resistance D,D-dipeptidase VanXYg | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D,D-dipeptidase/D,D-carboxypeptidase, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Evdokimova, E, Minasov, G, Egorova, O, Di Leo, R, Kudritska, M, Yim, V, Meziane-Cherif, D, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-15 | | Release date: | 2012-05-23 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1M25
 
 | | STRUCTURE OF SYNTHETIC 26-MER PEPTIDE CONTAINING 145-169 SHEEP PRION PROTEIN SEGMENT AND C-TERMINAL CYSTEINE IN TFE SOLUTION | | Descriptor: | MAJOR PRION PROTEIN | | Authors: | Megy, S, Bertho, G, Kozin, S.A, Coadou, G, Debey, P, Hoa, G.H, Girault, J.-P. | | Deposit date: | 2002-06-21 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Possible role of region 152-156 in the structural duality of a peptide fragment from sheep prion protein
Protein Sci., 13, 2004
|
|
1VHB
 
 | | BACTERIAL DIMERIC HEMOGLOBIN FROM VITREOSCILLA STERCORARIA | | Descriptor: | HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tarricone, C, Galizzi, A, Coda, A, Ascenzi, P, Bolognesi, M. | | Deposit date: | 1997-02-19 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Unusual structure of the oxygen-binding site in the dimeric bacterial hemoglobin from Vitreoscilla sp.
Structure, 5, 1997
|
|
1VIW
 
 | | TENEBRIO MOLITOR ALPHA-AMYLASE-INHIBITOR COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-AMYLASE, ALPHA-AMYLASE-INHIBITOR, ... | | Authors: | Nahoum, V, Egloff, M.P, Payan, F. | | Deposit date: | 1998-07-21 | | Release date: | 1999-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A plant-seed inhibitor of two classes of alpha-amylases: X-ray analysis of Tenebrio molitor larvae alpha-amylase in complex with the bean Phaseolus vulgaris inhibitor.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4F2F
 
 | | Crystal structure of the metal binding domain (MBD) of the Streptococcus pneumoniae D39 Cu(I) exporting P-type ATPase CopA with Cu(I) | | Descriptor: | CHLORIDE ION, COPPER (I) ION, Cation-transporting ATPase, ... | | Authors: | Fu, Y, Dann III, C.E, Giedroc, D.P. | | Deposit date: | 2012-05-07 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A new structural paradigm in copper resistance in Streptococcus pneumoniae.
Nat.Chem.Biol., 9, 2013
|
|
4FGN
 
 | |
4O5W
 
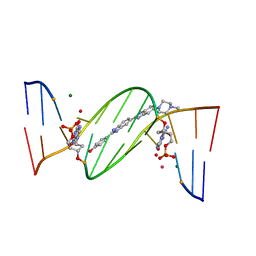 | | O6-carboxymethylguanine in DNA forms a sequence context dependent wobble base pair structure with thymine | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(C6G)P*AP*AP*TP*TP*TP*GP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Zhang, F, Tsunoda, M, Suzuki, K, Kikuchi, Y, Wilkinson, O, Millington, C.L, Margison, G.P, Williams, D.M, Takenaka, A. | | Deposit date: | 2013-12-20 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | O(6)-Carboxymethylguanine in DNA forms a sequence context-dependent wobble base-pair structure with thymine
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O5Z
 
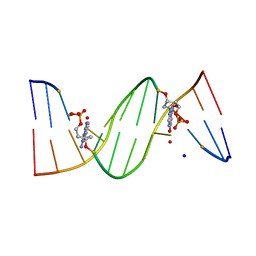 | | O6-carboxymethylguanine in DNA forms a sequence context dependent wobble base pair structure with thymine | | Descriptor: | BARIUM ION, DNA (5'-D(*CP*GP*CP*(C6G)P*AP*AP*TP*TP*TP*GP*CP*G)-3'), SODIUM ION | | Authors: | Zhang, F, Tsunoda, M, Suzuki, K, Kikuchi, Y, Wilkinson, O, Millington, C.L, Margison, G.P, Williams, D.M, Takenaka, A. | | Deposit date: | 2013-12-20 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | O(6)-Carboxymethylguanine in DNA forms a sequence context-dependent wobble base-pair structure with thymine
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8SJS
 
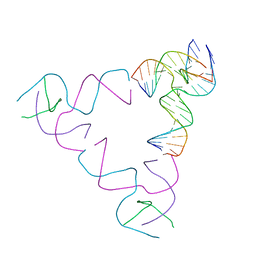 | | [3T18] Self-assembling right-handed tensegrity triangle with 18 interjunction base pairs and P63 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*GP*CP*CP*TP*GP*AP*CP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*CP*TP*GP*TP*GP*GP*CP*TP*C)-3'), DNA (5'-D(P*TP*CP*GP*TP*GP*GP*AP*CP*AP*GP*CP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (6.31 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4UYG
 
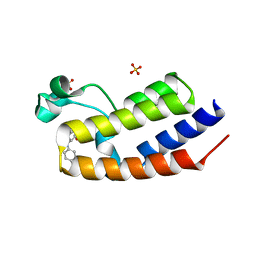 | | C-Terminal bromodomain of Human BRD2 with I-BET726 (GSK1324726A) | | Descriptor: | 4-[(2S,4R)-1-acetyl-4-[(4-chlorophenyl)amino]-2-methyl-1,2,3,4-tetrahydroquinolin-6-yl]benzoic acid, BROMODOMAIN-CONTAINING PROTEIN 2, SULFATE ION | | Authors: | Chung, C, Bamborough, P, Gosmini, R. | | Deposit date: | 2014-08-31 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Discovery of I-Bet726 (Gsk1324726A), a Potent Tetrahydroquinoline Apoa1 Up-Regulator and Selective Bet Bromodomain Inhibitor.
J.Med.Chem., 57, 2014
|
|
4FEU
 
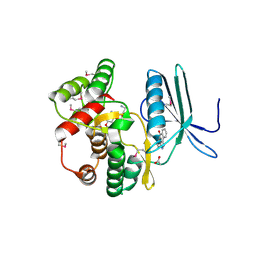 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor anthrapyrazolone SP600125 | | Descriptor: | 2,6-DIHYDROANTHRA/1,9-CD/PYRAZOL-6-ONE, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
1W20
 
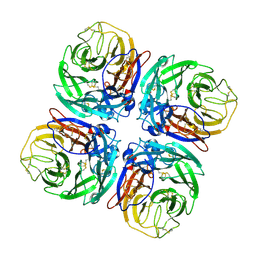 | | Structure of Neuraminidase from English duck subtype N6 complexed with 30 mM sialic acid (NANA, Neu5Ac), crystal soaked for 3 hours at 291 K | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Rudino-Pinera, E, Tunnah, P, Crennell, S.J, Webster, R.G, Laver, W.G, Garman, E.F. | | Deposit date: | 2004-06-24 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The Crystal Structure of Type a Influenza Virus Neuraminidase of the N6 Subtype Reveals the Existence of Two Separate Neu5Ac Binding Sites
To be Published
|
|
1W2W
 
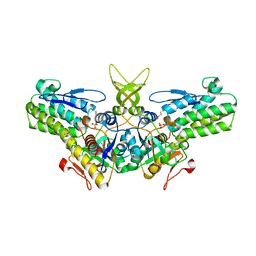 | | Crystal structure of yeast Ypr118w, a methylthioribose-1-phosphate isomerase related to regulatory eIF2B subunits | | Descriptor: | 5-METHYLTHIORIBOSE-1-PHOSPHATE ISOMERASE, SULFATE ION | | Authors: | Bumann, M, Djafarzadeh, S, Oberholzer, A.E, Bigler, P, Altmann, M, Trachsel, H, Baumann, U. | | Deposit date: | 2004-07-09 | | Release date: | 2004-07-16 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Yeast Ypr118W, a Methylthioribose-1-Phosphate Isomerase Related to Regulatory Eif2B Subunits
J.Biol.Chem., 279, 2004
|
|
4UYE
 
 | | BROMODOMAIN OF HUMAN BRPF1 WITH N-1,3-dimethyl-2-oxo-6-(piperidin-1- yl)-2,3-dihydro-1H-1,3-benzodiazol-5-yl-2-methoxybenzamide | | Descriptor: | 1,2-ETHANEDIOL, N-[1,3-dimethyl-2-oxo-6-(piperidin-1-yl)-2,3-dihydro-1H-benzimidazol-5-yl]-2-methoxybenzamide, PEREGRIN | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2014-08-30 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 1,3-Dimethyl Benzimidazolones are Potent, Selective Inhibitors of the Brpf1 Bromodomain.
Acs Med.Chem.Lett., 5, 2014
|
|
5D51
 
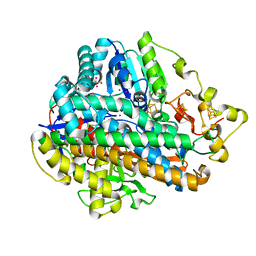 | | Krypton derivatization of an O2-tolerant membrane-bound [NiFe] hydrogenase reveals a hydrophobic gas tunnel network | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Kalms, J, Schmidt, A, Frielingsdorf, S, van der Linden, P, von Stetten, D, Lenz, O, Carpentier, P, Scheerer, P. | | Deposit date: | 2015-08-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Krypton Derivatization of an O2 -Tolerant Membrane-Bound [NiFe] Hydrogenase Reveals a Hydrophobic Tunnel Network for Gas Transport.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
