3VHP
 
 | | The insertion mutant Y61GG of Tm Cel12A | | 分子名称: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Cheng, Y.-S, Ko, T.-P, Guo, R.-T, Liu, J.-R. | | 登録日 | 2011-08-30 | | 公開日 | 2012-07-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Enhanced activity of Thermotoga maritima cellulase 12A by mutating a unique surface loop
Appl.Microbiol.Biotechnol., 95, 2012
|
|
8C3C
 
 | |
6DR8
 
 | | Metallo-beta-lactamase from Cronobacter sakazakii (Enterobacter sakazakii) HARLDQ motif mutant S60/R118H/Q121H/K254H | | 分子名称: | (2-hydroxyethoxy)acetaldehyde, Beta-lactamase, PHOSPHATE ION, ... | | 著者 | Monteiro Pedroso, M, Waite, D, Natasa, M, McGeary, R, Guddat, L, Hugenholtz, P, Schenk, G. | | 登録日 | 2018-06-11 | | 公開日 | 2019-06-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.476 Å) | | 主引用文献 | Broad spectrum antibiotic-degrading metallo-beta-lactamases are phylogenetically diverse
Protein Cell, 2020
|
|
8CO6
 
 | |
6DRJ
 
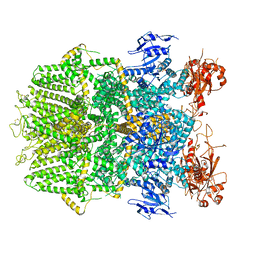 | | Structure of TRPM2 ion channel receptor by single particle electron cryo-microscopy, ADPR/Ca2+ bound state | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, CALCIUM ION, Transient receptor potential cation channel, ... | | 著者 | Du, J, Lu, W, Huang, Y, Winkler, P, Sun, W. | | 登録日 | 2018-06-12 | | 公開日 | 2018-09-19 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Architecture of the TRPM2 channel and its activation mechanism by ADP-ribose and calcium.
Nature, 562, 2018
|
|
3QGV
 
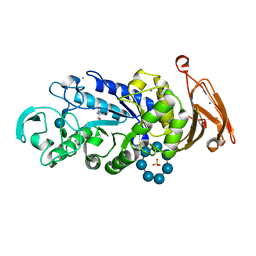 | | Crystal structure of a thermostable amylase variant | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha amylase, CALCIUM ION, ... | | 著者 | Hein, K.L, Ganshaw, G, Bott, R, Nissen, P. | | 登録日 | 2011-01-25 | | 公開日 | 2012-02-01 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of a highly mutated, thermostable xCExB1-amylase variant
To be Published
|
|
2ME0
 
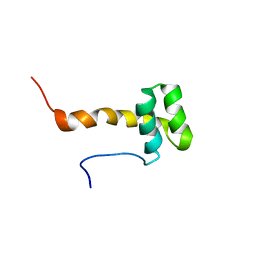 | |
1OKB
 
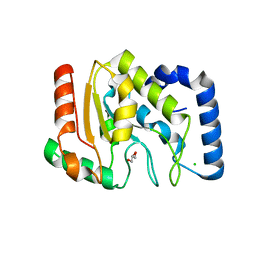 | | crystal structure of Uracil-DNA glycosylase from Atlantic cod (Gadus morhua) | | 分子名称: | CHLORIDE ION, GLYCEROL, URACIL-DNA GLYCOSYLASE | | 著者 | Leiros, I, Moe, E, Lanes, O, Smalas, A.O, Willassen, N.P. | | 登録日 | 2003-07-21 | | 公開日 | 2004-04-05 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The Crystal Structure of Uracil-DNA Glycosylase from Atlantic Cod (Gadus Morhua) Reveals Cold-Adaptation Features
Acta Crystallogr.,Sect.D, 59, 2003
|
|
7T4D
 
 | | Pore structure of pore-forming toxin Epx4 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Epx4 | | 著者 | Xiong, X.Z, Dong, M, Yang, P, Abraham, J. | | 登録日 | 2021-12-09 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Emerging enterococcus pore-forming toxins with MHC/HLA-I as receptors.
Cell, 185, 2022
|
|
3VNI
 
 | | Crystal structures of D-Psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars | | 分子名称: | MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | 著者 | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | 登録日 | 2012-01-16 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
1OPP
 
 | | PEPTIDE OF HUMAN APOLIPOPROTEIN C-I RESIDUES 1-38, NMR, 28 STRUCTURES | | 分子名称: | APOLIPOPROTEIN C-I | | 著者 | Rozek, A, Buchko, G.W, Kanda, P, Cushley, R.J. | | 登録日 | 1997-05-08 | | 公開日 | 1998-05-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Conformational studies of the N-terminal lipid-associating domain of human apolipoprotein C-I by CD and 1H NMR spectroscopy.
Protein Sci., 6, 1997
|
|
7T4S
 
 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with NRP2 and neutralizing fabs 8I21 and 13H11 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein H, ... | | 著者 | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | 登録日 | 2021-12-10 | | 公開日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
7T4Q
 
 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with neutralizing fabs 2C12, 7I13 and 13H11 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | 著者 | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | 登録日 | 2021-12-10 | | 公開日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
1P4I
 
 | | Crystal Structure of scFv against peptide GCN4 | | 分子名称: | ANTIBODY VARIABLE LIGHT CHAIN, antibody variable heavy chain | | 著者 | Zahnd, C, Spinelli, S, Luginbuhl, B, Jermutus, L, Amstutz, P, Cambillau, C, Pluckthun, A. | | 登録日 | 2003-04-23 | | 公開日 | 2004-05-04 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Directed in Vitro Evolution and Crystallographic Analysis of a Peptide-binding Single Chain Antibody Fragment (scFv) with Low Picomolar Affinity.
J.Biol.Chem., 279, 2004
|
|
8CJ2
 
 | | Urea-based foldamer inhibitor c3u_5 chimera in complex with ASF1 histone chaperone | | 分子名称: | GLYCEROL, Histone chaperone ASF1A, SULFATE ION, ... | | 著者 | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | 登録日 | 2023-02-11 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.127 Å) | | 主引用文献 | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
3VED
 
 | | Rhodococcus jostii RHA1 DypB D153H variant in complex with heme | | 分子名称: | CHLORIDE ION, DypB, GLYCEROL, ... | | 著者 | Grigg, J.C, Singh, R, Armstrong, Z, Eltis, L.D, Murphy, M.E.P. | | 登録日 | 2012-01-07 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Distal heme pocket residues of B-type dye-decolorizing peroxidase: arginine but not aspartate is essential for peroxidase activity.
J.Biol.Chem., 287, 2012
|
|
7KE8
 
 | |
8CJ1
 
 | | Urea-based foldamer inhibitor c3u_3 chimera in complex with ASF1 histone chaperone | | 分子名称: | Histone chaperone ASF1A, c3u_3 chimera inhibitor of histone chaperone ASF1 | | 著者 | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | 登録日 | 2023-02-11 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.564 Å) | | 主引用文献 | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
6NZG
 
 | | Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine | | 分子名称: | (1S,2R,3S,4S,5S,6R)-2-amino-3,4,5,6-tetrahydroxycyclohexane-1-carboxylic acid, Beta-galactosidase, CALCIUM ION, ... | | 著者 | Pellock, S.J, Jariwala, P.B, Redinbo, M.R. | | 登録日 | 2019-02-13 | | 公開日 | 2019-12-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Discovering the Microbial Enzymes Driving Drug Toxicity with Activity-Based Protein Profiling.
Acs Chem.Biol., 15, 2020
|
|
3QP4
 
 | | Crystal structure of CviR ligand-binding domain bound to C10-HSL | | 分子名称: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]decanamide | | 著者 | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | 登録日 | 2011-02-11 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
6NDB
 
 | |
6NDH
 
 | |
7TF4
 
 | | Cryo-EM structure of SARS-CoV-2 Kappa (B.1.617.1) spike protein (focused refinement of RBD) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | 登録日 | 2022-01-06 | | 公開日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
1P9K
 
 | | THE SOLUTION STRUCTURE OF YBCJ FROM E. COLI REVEALS A RECENTLY DISCOVERED ALFAL MOTIF INVOLVED IN RNA-BINDING | | 分子名称: | orf, hypothetical protein | | 著者 | Volpon, L, Lievre, C, Osborne, M.J, Gandhi, S, Iannuzzi, P, Larocque, R, Matte, A, Cygler, M, Gehring, K, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | 登録日 | 2003-05-12 | | 公開日 | 2003-11-25 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of YbcJ from Escherichia coli reveals a recently discovered alphaL motif involved in RNA binding.
J.Bacteriol., 185, 2003
|
|
6YQ3
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | 分子名称: | (3~{R})-8-methoxy-3-methyl-3,6-bis(oxidanyl)-2,4-dihydrobenzo[a]anthracene-1,7,12-trione, 1,2-ETHANEDIOL, Monooxygenase, ... | | 著者 | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | 登録日 | 2020-04-16 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
