6RNF
 
 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Xylose Isomerase 30 ms timepoint | | 分子名称: | MAGNESIUM ION, Xylose isomerase, alpha-D-glucopyranose | | 著者 | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | 登録日 | 2019-05-08 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
7VJU
 
 | |
7MFG
 
 | | Cryo-EM structure of the VRC310 clinical trial, vaccine-elicited, human antibody 310-030-1D06 Fab in complex with an H1 NC99 HA trimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 310-030-1D06 Heavy, 310-030-1D06 Light, ... | | 著者 | Gorman, J, Kwong, P.D. | | 登録日 | 2021-04-09 | | 公開日 | 2021-11-03 | | 最終更新日 | 2022-03-09 | | 実験手法 | ELECTRON MICROSCOPY (3.87 Å) | | 主引用文献 | A single residue in influenza virus H2 hemagglutinin enhances the breadth of the B cell response elicited by H2 vaccination.
Nat Med, 28, 2022
|
|
6IBD
 
 | | The Phosphatase and C2 domains of Human SHIP1 | | 分子名称: | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | 著者 | Bradshaw, W.J, Williams, E.P, Fernandez-Cid, A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2018-11-29 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
8CH8
 
 | | Crystal structure of the human PXR ligand-binding domain in complex with liranaftate | | 分子名称: | Nuclear receptor subfamily 1 group I member 2, ~{O}-(5,6,7,8-tetrahydronaphthalen-2-yl) ~{N}-(6-methoxypyridin-2-yl)-~{N}-methyl-carbamothioate | | 著者 | Carivenc, C, Derosa, Q, Grimaldi, M, Boulahtouf, A, Balaguer, P, Bourguet, W. | | 登録日 | 2023-02-07 | | 公開日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of the human PXR ligand-binding domain in complex with liranaftate
To Be Published
|
|
8CHD
 
 | | NtUGT1 in two conformations | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Glycosyltransferase, ... | | 著者 | Fredslund, F, Teze, D, Adams, P.D, Welner, D.H. | | 登録日 | 2023-02-07 | | 公開日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | NtUGT1 in two conformations
To Be Published
|
|
7MX4
 
 | | CD1c with antigen analogue 1 | | 分子名称: | (2S)-2,3-dihydroxypropyl hexadecanoate, (4S,8R,12S,16S,20S)-4,8,12,16,20-pentamethylheptacosyl (1R,2S,3S,4R,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl hydrogen (R)-phosphate, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | 著者 | Cao, T.P, Shahine, A, Rossjohn, J. | | 登録日 | 2021-05-18 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Rational design of a hydrolysis-resistant mycobacterial phosphoglycolipid antigen presented by CD1c to T cells.
J.Biol.Chem., 297, 2021
|
|
8CID
 
 | |
7M7I
 
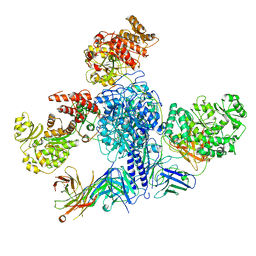 | | 6-Deoxyerythronolide B synthase (DEBS) module 1 in complex with antibody fragment 1B2 (TE-free) | | 分子名称: | 1B2 (heavy chain), 1B2 (light chain), EryAI, ... | | 著者 | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | 登録日 | 2021-03-28 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
6RSE
 
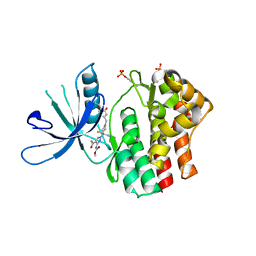 | |
6WQB
 
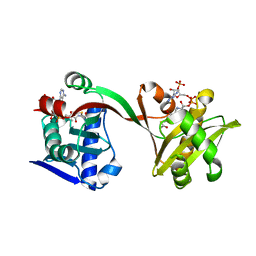 | | Crystal structure of VipF from Legionella hackeliae in complex with acetyl-CoA | | 分子名称: | ACETYL COENZYME *A, N-terminal acetyltransferase, GNAT family | | 著者 | Stogios, P.J, Skarina, T, Wawrzak, Z, Sandoval, J, Di Leo, R, Savchenko, A. | | 登録日 | 2020-04-28 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of VipF from Legionella hackeliae in complex with acetyl-CoA
To Be Published
|
|
6WQC
 
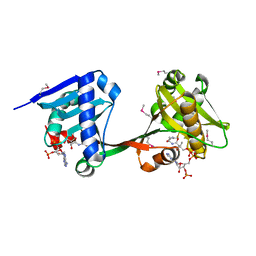 | | Crystal structure of VipF from Legionella hackeliae in complex with CoA | | 分子名称: | COENZYME A, N-terminal acetyltransferase, GNAT family | | 著者 | Stogios, P.J, Skarina, T, Wawrzak, Z, Sandoval, J, Di Leo, R, Savchenko, A. | | 登録日 | 2020-04-28 | | 公開日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Crystal structure of VipF from Legionella hackeliae in complex with CoA
To Be Published
|
|
1OIR
 
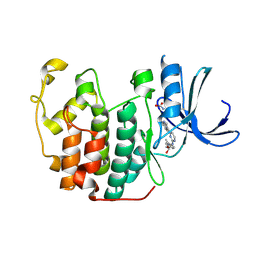 | | Imidazopyridines: a potent and selective class of Cyclin-dependent Kinase inhibitors identified through Structure-based hybridisation | | 分子名称: | 1-(DIMETHYLAMINO)-3-(4-{{4-(2-METHYLIMIDAZO[1,2-A]PYRIDIN-3-YL)PYRIMIDIN-2-YL]AMINO}PHENOXY)PROPAN-2-OL, CELL DIVISION PROTEIN KINASE 2 | | 著者 | Beattie, J.F, Breault, G.A, Byth, K.F, Culshaw, J.D, Ellston, R.P.A, Green, S, Minshull, C.A, Norman, R.A, Pauptit, R.A, Thomas, A.P, Jewsbury, P.J. | | 登録日 | 2003-06-24 | | 公開日 | 2003-09-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Imidazo[1,2-A]Pyridines: A Potent and Selective Class of Cyclin-Dependent Kinase Inhibitors Identified Through Structure-Based Hybridisation
Bioorg.Med.Chem.Lett., 13, 2003
|
|
7VY2
 
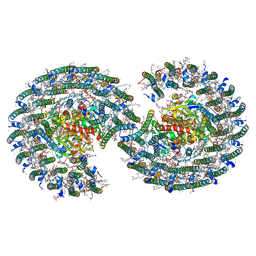 | | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF RHODOBACTER SPHAEROIDES DIMER | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Antenna pigment protein alpha chain, Antenna pigment protein beta chain, ... | | 著者 | Tani, K, Kanno, R, Kawamura, S, Kikuchi, R, Nagashima, K.V.P, Hall, M, Takahashi, A, Yu, L.-J, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | 登録日 | 2021-11-13 | | 公開日 | 2022-04-27 | | 実験手法 | ELECTRON MICROSCOPY (2.75 Å) | | 主引用文献 | Asymmetric structure of the native Rhodobacter sphaeroides dimeric LH1-RC complex.
Nat Commun, 13, 2022
|
|
7M7F
 
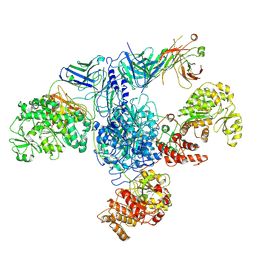 | | 6-Deoxyerythronolide B synthase (DEBS) module 1 in complex with antibody fragment 1B2: State 1 | | 分子名称: | 1B2 (heavy chain), 1B2 (light chain), EryAI,6-deoxyerythronolide-B synthase EryA3, ... | | 著者 | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | 登録日 | 2021-03-28 | | 公開日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
5DXH
 
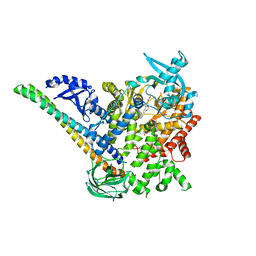 | | p110alpha/p85alpha with compound 5 | | 分子名称: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, methyl {2-[4-(2-chlorophenyl)-4H-1,2,4-triazol-3-yl]-4,5-dihydrothieno[3,2-d][1]benzoxepin-8-yl}carbamate | | 著者 | Heffron, T.P, Heald, R.A, Ndubaku, C, Wei, B.Q, Augustin, M, Do, S, Edgar, K, Eigenbrot, C, Friedman, L, Gancia, E, Jackson, P.S, Jones, G, Kolesnikov, A, Lee, L.B, Lesnick, J.D, Lewis, C, McLean, N, Mortle, M, Nonomiya, J, Pang, J, Price, S, Prior, W.W, Salphati, L, Sideris, S, Staben, S, Steinbacher, S, Tsui, V, Wallin, J, Sampath, D, Olivero, A. | | 登録日 | 2015-09-23 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The Rational Design of Selective Benzoxazepin Inhibitors of the alpha-Isoform of Phosphoinositide 3-Kinase Culminating in the Identification of (S)-2-((2-(1-Isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepin-9-yl)oxy)propanamide (GDC-0326).
J.Med.Chem., 59, 2016
|
|
6I23
 
 | | Flavin Analogue Sheds Light on Light-Oxygen-Voltage Domain Mechanism | | 分子名称: | 1,2-ETHANEDIOL, 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-5-O-phosphono-D-ribitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Rizkallah, P.J, Kalvaitis, M.E, Allemann, R.K, Mart, R.J, Johnson, L.A. | | 登録日 | 2018-10-31 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Noncanonical Chromophore Reveals Structural Rearrangements of the Light-Oxygen-Voltage Domain upon Photoactivation.
Biochemistry, 58, 2019
|
|
6XF7
 
 | | SLP | | 分子名称: | Lambda 1 protein | | 著者 | Sutton, G, Sun, D.P, Fu, X.F, Kotecha, A, Hecksel, G.W, Clare, D.K, Zhang, P, Stuart, D, Boyce, M. | | 登録日 | 2020-06-15 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (6.6 Å) | | 主引用文献 | Assembly intermediates of orthoreovirus captured in the cell.
Nat Commun, 11, 2020
|
|
1ENM
 
 | | UDA TRISACCHARIDE COMPLEX. CRYSTAL STRUCTURE OF URTICA DIOICA AGGLUTININ, A SUPERANTIGEN PRESENTED BY MHC MOLECULES OF CLASS I AND CLASS II | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AGGLUTININ ISOLECTIN I/AGGLUTININ ISOLECTIN V/ AGGLUTININ ISOLECTIN VI | | 著者 | Saul, F.A, Rovira, P, Boulot, G, Van Damme, E.J.M, Peumans, W.J, Truffa-Bachi, P, Bentley, G.A. | | 登録日 | 2000-03-21 | | 公開日 | 2000-06-21 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of Urtica dioica agglutinin, a superantigen presented by MHC molecules of class I and class II.
Structure Fold.Des., 8, 2000
|
|
6S0S
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel, ribostamycin B and 2-oxoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, CHLORIDE ION, Kanamycin B dioxygenase, ... | | 著者 | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Minor, W, Borowski, T. | | 登録日 | 2019-06-18 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
8H10
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8CSA
 
 | |
7M74
 
 | | ATP-bound AMP-activated protein kinase | | 分子名称: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | 著者 | Yan, Y, Mukherjee, S, Harikumar, K.G, Strutzenberg, T, Zhou, X.E, Powell, S.K, Xu, T, Sheldon, R, Lamp, J, Brunzelle, J.S, Radziwon, K, Ellis, A, Novick, S.J, Vega, I.E, Jones, R, Miller, L.J, Xu, H.E, Griffin, P.R, Kossiakoff, A.A, Melcher, K. | | 登録日 | 2021-03-26 | | 公開日 | 2021-12-15 | | 最終更新日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
8H13
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H14
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x3 Disulfide (D414C and V969C), Locked-1 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
