3ERI
 
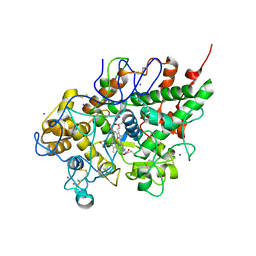 | | First structural evidence of substrate specificity in mammalian peroxidases: Crystal structures of substrate complexes with lactoperoxidases from two different species | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Singh, A.K, Singh, N, Sheikh, I.A, Sinha, M, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | 登録日 | 2008-10-02 | | 公開日 | 2009-03-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Evidence of Substrate Specificity in Mammalian Peroxidases: STRUCTURE OF THE THIOCYANATE COMPLEX WITH LACTOPEROXIDASE AND ITS INTERACTIONS AT 2.4 A RESOLUTION
J.Biol.Chem., 284, 2009
|
|
8Q91
 
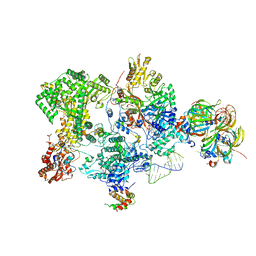 | | Structure of the human 20S U5 snRNP core | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Schneider, S, Galej, W.P. | | 登録日 | 2023-08-19 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure of the human 20S U5 snRNP.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7VH5
 
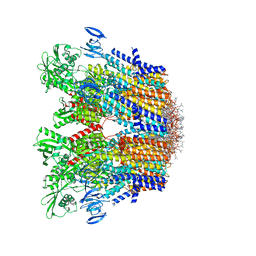 | | Cryo-EM structure of the hexameric plasma membrane H+-ATPase in the autoinhibited state (pH 7.4, C1 symmetry) | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Plasma membrane ATPase 1, SPHINGOSINE | | 著者 | Zhao, P, Zhao, C, Chen, D, Yun, C, Li, H, Bai, L. | | 登録日 | 2021-09-21 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure and activation mechanism of the hexameric plasma membrane H + -ATPase.
Nat Commun, 12, 2021
|
|
7VH6
 
 | | Cryo-EM structure of the hexameric plasma membrane H+-ATPase in the active state (pH 6.0, BeF3-, conformation 1, C1 symmetry) | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, BERYLLIUM TRIFLUORIDE ION, Plasma membrane ATPase 1 | | 著者 | Zhao, P, Zhao, C, Chen, D, Yun, C, Li, H, Bai, L. | | 登録日 | 2021-09-21 | | 公開日 | 2021-11-24 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure and activation mechanism of the hexameric plasma membrane H + -ATPase.
Nat Commun, 12, 2021
|
|
5JUU
 
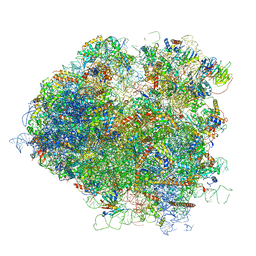 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure V (least rotated 40S subunit) | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | 著者 | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | 登録日 | 2016-05-10 | | 公開日 | 2016-10-05 | | 最終更新日 | 2019-11-27 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
3EAM
 
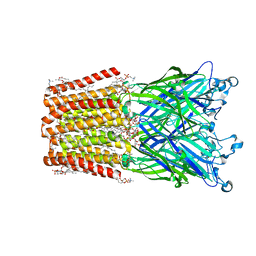 | | An open-pore structure of a bacterial pentameric ligand-gated ion channel | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | 著者 | Bocquet, N, Nury, H, Baaden, M, Le Poupon, C, Changeux, J.P, Delarue, M, Corringer, P.J. | | 登録日 | 2008-08-26 | | 公開日 | 2008-11-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation.
Nature, 457, 2009
|
|
7SYT
 
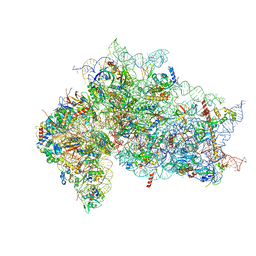 | | Structure of the wt IRES w/o eIF2 48S initiation complex, closed conformation. Structure 13(wt) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
5K9J
 
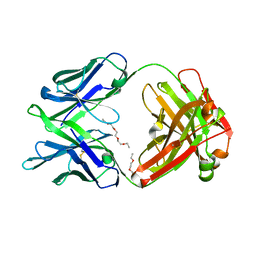 | | Crystal structure of multidonor HV6-1-class broadly neutralizing Influenza A antibody 56.a.09 isolated following H5 immunization. | | 分子名称: | 56.a.09 heavy chain, 56.a.09 light chain, POLYETHYLENE GLYCOL (N=34) | | 著者 | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | 登録日 | 2016-05-31 | | 公開日 | 2016-07-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.904 Å) | | 主引用文献 | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell, 166, 2016
|
|
3EMZ
 
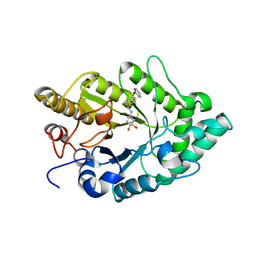 | | Crystal structure of xylanase XynB from Paenibacillus barcinonensis complexed with a conduramine derivative | | 分子名称: | (1S,2S,3R,6R)-6-[(4-phenoxybenzyl)amino]cyclohex-4-ene-1,2,3-triol, Endo-1,4-beta-xylanase | | 著者 | Sanz-Aparicio, J, Isorna, P. | | 登録日 | 2008-09-25 | | 公開日 | 2009-09-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structural insights into the specificity of Xyn10B from Paenibacillus barcinonensis and its improved stability by forced protein evolution.
J.Biol.Chem., 285, 2010
|
|
5JUS
 
 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure III (mid-rotated 40S subunit) | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | 著者 | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | 登録日 | 2016-05-10 | | 公開日 | 2016-10-05 | | 最終更新日 | 2023-04-05 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
7SYP
 
 | | Structure of the wt IRES and 40S ribosome binary complex, open conformation. Structure 10(wt) | | 分子名称: | 18S rRNA, HCV IRES, HCV IRES partially loaded mRNA portion, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-27 | | 最終更新日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYO
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head open. Structure 9(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, HCV IRES, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-27 | | 最終更新日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYS
 
 | | Structure of the delta dII IRES eIF2-containing 48S initiation complex, closed conformation. Structure 12(delta dII). | | 分子名称: | 18S rRNA, Eukaryotic translation initiation factor 1A, X-chromosomal, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-27 | | 最終更新日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYQ
 
 | | Structure of the wt IRES and 40S ribosome ternary complex, open conformation. Structure 11(wt) | | 分子名称: | 18S rRNA, Eukaryotic translation initiation factor 1A, X-chromosomal, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-27 | | 最終更新日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
8VW4
 
 | | Crystal structure of Cbl-b TKB bound to compound 26 | | 分子名称: | (7-methoxy-2-{2-[(1S,3S,4S)-3-(3-methoxy-2-methyl-5-nitrophenyl)-1-methyl-5-oxo-1,5-dihydroimidazo[1,5-a]pyridin-2(3H)-yl]-2-oxoethoxy}quinolin-8-yl)acetic acid, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase CBL-B, ... | | 著者 | Yu, C, Murray, J, Hsu, P.L. | | 登録日 | 2024-01-31 | | 公開日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Optimization of a Novel DEL Hit That Binds in the Cbl-b SH2 Domain and Blocks Substrate Binding.
Acs Med.Chem.Lett., 15, 2024
|
|
7SYR
 
 | | Structure of the wt IRES eIF2-containing 48S initiation complex, closed conformation. Structure 12(wt). | | 分子名称: | 18S rRNA, Eukaryotic translation initiation factor 1A, X-chromosomal, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-27 | | 最終更新日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
3E70
 
 | | Structures and conformations in solution of the Signal Recognition Particle Receptor from the Archaeon Pyrococcus Furiosus | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Signal recognition particle receptor | | 著者 | Egea, P.F, Tsuruta, H, Napetschnig, J, Walter, P, Stroud, R.M. | | 登録日 | 2008-08-17 | | 公開日 | 2008-11-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structures of the signal recognition particle receptor from the archaeon Pyrococcus furiosus: implications for the targeting step at the membrane.
Plos One, 3, 2008
|
|
8B5S
 
 | | Crystal Structure of P. aeruginosa WaaG in complex with UDP-glucose | | 分子名称: | UDP-glucose:(Heptosyl) LPS alpha 1,3-glucosyltransferase WaaG, URIDINE-5'-DIPHOSPHATE, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | 著者 | Scaletti, E, Gustafsson Westergren, R, Stenmark, P. | | 登録日 | 2022-09-24 | | 公開日 | 2023-10-04 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
J.Biol.Chem., 299, 2023
|
|
8B5Q
 
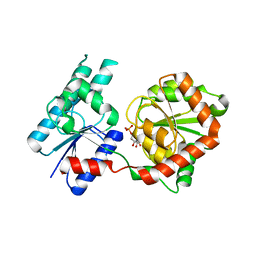 | |
8B62
 
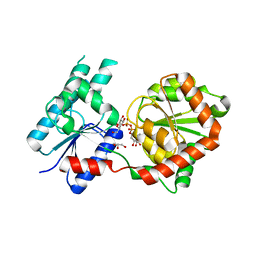 | | Crystal Structure of P. aeruginosa WaaG in complex with UDP-galactose | | 分子名称: | GALACTOSE-URIDINE-5'-DIPHOSPHATE, GLYCEROL, UDP-glucose:(Heptosyl) LPS alpha 1,3-glucosyltransferase WaaG | | 著者 | Scaletti, E, Gustafsson Westergren, R, Stenmark, P. | | 登録日 | 2022-09-25 | | 公開日 | 2023-10-04 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
J.Biol.Chem., 299, 2023
|
|
8B63
 
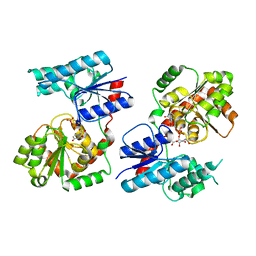 | | Crystal Structure of P. aeruginosa WaaG in complex with UDP-GalNAc | | 分子名称: | ACETATE ION, UDP-glucose:(Heptosyl) LPS alpha 1,3-glucosyltransferase WaaG, URIDINE-5'-DIPHOSPHATE, ... | | 著者 | Scaletti, E, Gustafsson Westergren, R, Stenmark, P. | | 登録日 | 2022-09-25 | | 公開日 | 2023-10-04 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
J.Biol.Chem., 299, 2023
|
|
6RNY
 
 | | PFV intasome - nucleosome strand transfer complex | | 分子名称: | DNA (108-MER), DNA (128-MER), DNA (33-MER), ... | | 著者 | Pye, V.E, Renault, L, Maskell, D.P, Cherepanov, P, Costa, A. | | 登録日 | 2019-05-09 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Retroviral integration into nucleosomes through DNA looping and sliding along the histone octamer.
Nat Commun, 10, 2019
|
|
8B7P
 
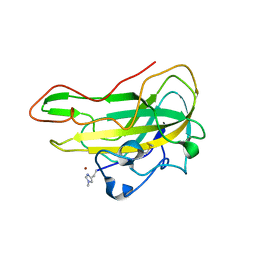 | | Crystal structure of an AA9 LPMO from Aspergillus nidulans, AnLPMOC | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Endo-beta-1,4-glucanase D | | 著者 | Males, A, Rafael Fanchini Terrasan, C, Davies, G.J, Walton, P.H. | | 登録日 | 2022-09-30 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Characterisation of lytic polysaccharide monooxygenases from Aspergillus nidulans
To Be Published
|
|
5LZL
 
 | | Pyrobaculum calidifontis 5-aminolaevulinic acid dehydratase | | 分子名称: | Delta-aminolevulinic acid dehydratase, ZINC ION | | 著者 | Azim, N, Erskine, P.T, Guo, J, Cooper, J.B. | | 登録日 | 2016-09-30 | | 公開日 | 2016-10-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.47 Å) | | 主引用文献 | Structural studies of substrate and product complexes of 5-aminolaevulinic acid dehydratase from humans, Escherichia coli and the hyperthermophile Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6HCY
 
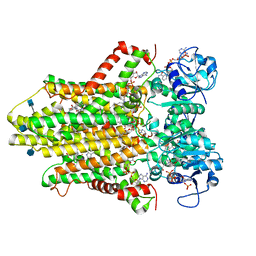 | | human STEAP4 bound to NADP, FAD, heme and Fe(III)-NTA. | | 分子名称: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Oosterheert, W, van Bezouwen, L.S, Rodenburg, R.N.P, Forster, F, Mattevi, A, Gros, P. | | 登録日 | 2018-08-17 | | 公開日 | 2018-10-24 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cryo-EM structures of human STEAP4 reveal mechanism of iron(III) reduction.
Nat Commun, 9, 2018
|
|
