4X3J
 
 | | Selection of fragments for kinase inhibitor design: decoration is key | | 分子名称: | 1-[4-(4-amino-5-oxopyrido[2,3-d]pyrimidin-8(5H)-yl)phenyl]-3-[2-fluoro-5-(trifluoromethyl)phenyl]urea, Angiopoietin-1 receptor | | 著者 | Czodrowski, P, Hoelzemann, G, Barnickel, G, Greiner, H, Musil, D. | | 登録日 | 2014-11-30 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Selection of fragments for kinase inhibitor design: decoration is key.
J.Med.Chem., 58, 2015
|
|
4X4M
 
 | | Structure of FcgammaRI in complex with Fc reveals the importance of glycan recognition for high affinity IgG binding | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Lu, J, Sun, P.D. | | 登録日 | 2014-12-03 | | 公開日 | 2015-04-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.485 Å) | | 主引用文献 | Structure of Fc gamma RI in complex with Fc reveals the importance of glycan recognition for high-affinity IgG binding.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1B1X
 
 | |
4WSL
 
 | | Crystal structure of designed cPPR-polyC protein | | 分子名称: | Pentatricopeptide repeat protein | | 著者 | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | 登録日 | 2014-10-28 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4WWK
 
 | | Crystal structure of human TCR Alpha Chain-TRAV12-3, Beta Chain-TRBV6-5, Antigen-presenting molecule CD1d, and Beta-2-microglobulin | | 分子名称: | (15Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]tetracos-15-enamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, ... | | 著者 | Le Nours, J, Praveena, T, Pellicci, D.G, Gherardin, N.A, Lim, R.T, Besra, G, Keshipeddy, A, Richardson, S.K, Howell, A.R, Gras, S, Godfrey, D.I, Rossjohn, J, Uldrich, A.P. | | 登録日 | 2014-11-11 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Atypical natural killer T-cell receptor recognition of CD1d-lipid antigens.
Nat Commun, 7, 2016
|
|
6WIG
 
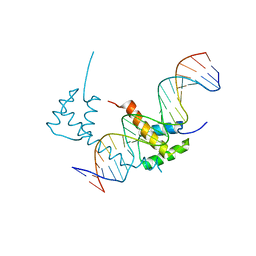 | | Structure of STENOFOLIA Protein HD domain bound with DNA | | 分子名称: | DNA (5'-D(P*CP*TP*TP*GP*AP*AP*TP*AP*AP*AP*TP*CP*AP*TP*TP*AP*AP*TP*TP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*AP*AP*TP*TP*AP*AP*TP*GP*AP*TP*TP*TP*AP*TP*TP*CP*AP*AP*G)-3'), STENOFOLIA | | 著者 | Deng, J, Peng, S, Pathak, P. | | 登録日 | 2020-04-09 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the unique tetrameric STENOFOLIA homeodomain bound with target promoter DNA.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4WUY
 
 | | Crystal Structure of Protein Lysine Methyltransferase SMYD2 in complex with LLY-507, a Cell-Active, Potent and Selective Inhibitor | | 分子名称: | 5-cyano-2'-{4-[2-(3-methyl-1H-indol-1-yl)ethyl]piperazin-1-yl}-N-[3-(pyrrolidin-1-yl)propyl]biphenyl-3-carboxamide, GLYCEROL, N-lysine methyltransferase SMYD2, ... | | 著者 | Nguyen, H, Allali-Hassani, A, Antonysamy, S, Chang, S, Chen, L.H, Curtis, C, Emtage, S, Fan, L, Gheyi, T, Li, F, Liu, S, Martin, J.R, Mendel, D, Olsen, J.B, Pelletier, L, Shatseva, T, Wu, S, Zhang, F.F, Arrowsmith, C.H, Brown, P.J, Campbell, R.M, Garcia, B.A, Barsyte-Lovejoy, D, Mader, M, Vedadi, M. | | 登録日 | 2014-11-04 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | LLY-507, a Cell-active, Potent, and Selective Inhibitor of Protein-lysine Methyltransferase SMYD2.
J.Biol.Chem., 290, 2015
|
|
4WW1
 
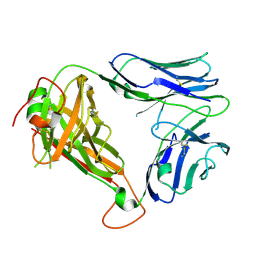 | | Crystal structure of human TCR Alpha Chain-TRAV21-TRAJ8 and Beta Chain-TRBV7-8 | | 分子名称: | TCR Alpha Chain-TRAV21-TRAJ8, TCR Beta Chain-TRBV7-8 | | 著者 | Le Nours, J, Praveena, T, Pellicci, D.G, Lim, R.T, Besra, G, Howell, A.R, Godfrey, D.I, Rossjohn, J, Uldrich, A.P. | | 登録日 | 2014-11-10 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Atypical natural killer T-cell receptor recognition of CD1d-lipid antigens.
Nat Commun, 7, 2016
|
|
4X0L
 
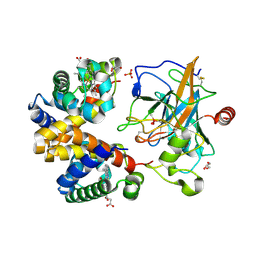 | | Human haptoglobin-haemoglobin complex | | 分子名称: | CACODYLATE ION, GLYCEROL, Haptoglobin, ... | | 著者 | Lane-Serff, H, MacGregor, P, Lowe, E.D, Carrington, M, Higgins, M.K. | | 登録日 | 2014-11-21 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural basis for ligand and innate immunity factor uptake by the trypanosome haptoglobin-haemoglobin receptor.
Elife, 3, 2014
|
|
4X0F
 
 | |
6ZH7
 
 | | Crystal structure of fatty acid photodecarboxylase in the dark state determined by serial femtosecond crystallography at room temperature | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | 著者 | Hadjidemetriou, K, Coquelle, N, Weik, M, Schlichting, I, Barends, T.R.M, Colletier, J.P. | | 登録日 | 2020-06-21 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
4X6I
 
 | | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors. | | 分子名称: | 2-amino-4-bromo-N-{1-[(cyanomethyl)carbamoyl]cyclohexyl}benzamide, Cathepsin K, SULFATE ION | | 著者 | Borisek, J, Mohar, B, Vizovisek, M, Sosnowski, P, Turk, D, Turk, B, Novic, M. | | 登録日 | 2014-12-08 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Development of N-(Functionalized benzoyl)-homocycloleucyl-glycinonitriles as Potent Cathepsin K Inhibitors.
J.Med.Chem., 58, 2015
|
|
6NM4
 
 | | Crystal structure of SAM-bound PRDM9 in complex with MRK-740 inhibitor | | 分子名称: | 4-[3-(3,5-dimethoxyphenyl)-1,2,4-oxadiazol-5-yl]-1-methyl-9-(2-methylpyridin-4-yl)-1,4,9-triazaspiro[5.5]undecane, Histone-lysine N-methyltransferase PRDM9, S-ADENOSYLMETHIONINE, ... | | 著者 | Ivanochko, D, Halabelian, L, Fischer, C, Sanders, J.M, Kattar, S.D, Brown, P.J, Edwards, A.M, Bountra, C, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | 登録日 | 2019-01-10 | | 公開日 | 2019-02-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Discovery of a chemical probe for PRDM9.
Nat Commun, 10, 2019
|
|
4WO8
 
 | | The substrate-free duplicated taurocyamine kinase from Schistosoma mansoni | | 分子名称: | Taurocyamine kinase | | 著者 | Merceron, R, Awama, A, Montserret, R, Marcillat, O, Gouet, P. | | 登録日 | 2014-10-15 | | 公開日 | 2015-04-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Substrate-free and -bound Crystal Structures of the Duplicated Taurocyamine Kinase from the Human Parasite Schistosoma mansoni.
J.Biol.Chem., 290, 2015
|
|
4X22
 
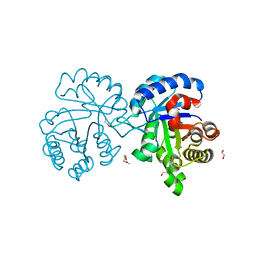 | | Crystal structure of Leptospira Interrogans Triosephosphate Isomerase (LiTIM) | | 分子名称: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Triosephosphate isomerase | | 著者 | Pareek, V, Balaram, P, Murthy, M.R.N. | | 登録日 | 2014-11-25 | | 公開日 | 2016-02-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.084 Å) | | 主引用文献 | Connecting Active-Site Loop Conformations and Catalysis in Triosephosphate Isomerase: Insights from a Rare Variation at Residue 96 in the Plasmodial Enzyme
Chembiochem, 17, 2016
|
|
4X2F
 
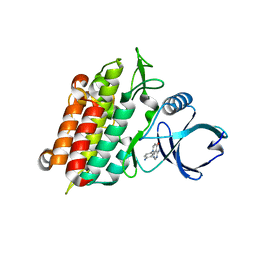 | | Selection of fragments for kinase inhibitor design: decoration is key | | 分子名称: | 4-amino-8-(4-aminophenyl)pyrido[2,3-d]pyrimidin-5(8H)-one, SULFATE ION, TGF-beta receptor type-1 | | 著者 | Czodrowski, P, Hoelzemann, G, Barnickel, G, Greiner, H, Musil, D. | | 登録日 | 2014-11-26 | | 公開日 | 2015-10-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Selection of fragments for kinase inhibitor design: decoration is key.
J.Med.Chem., 58, 2015
|
|
4X2N
 
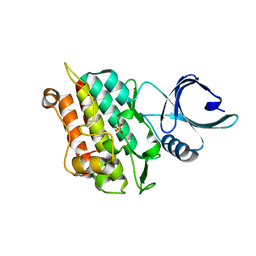 | | Selection of fragments for kinase inhibitor design: decoration is key | | 分子名称: | SULFATE ION, TGF-beta receptor type-1 | | 著者 | Czodrowski, P, Hoelzemann, G, Barnickel, G, Greiner, H, Musil, D. | | 登録日 | 2014-11-26 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Selection of fragments for kinase inhibitor design: decoration is key.
J.Med.Chem., 58, 2015
|
|
6XF6
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (1 RBD up) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-06-15 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-12-02 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
4WWD
 
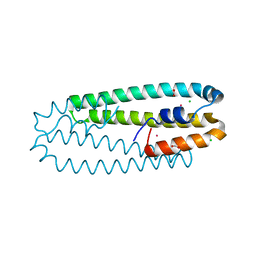 | | High-resolution structure of the Co-bound form of the Y135F mutant of C. metallidurans CnrXs | | 分子名称: | CARBON DIOXIDE, CHLORIDE ION, COBALT (II) ION, ... | | 著者 | Volbeda, A, Coves, J, Maillard, A.P, Kuennemann, S, Grosse, C, Schleuder, G, Petit-Haertlein, I, de Rosny, E, Nies, D.H. | | 登録日 | 2014-11-10 | | 公開日 | 2015-02-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Response of CnrX from Cupriavidus metallidurans CH34 to nickel binding.
Metallomics, 7, 2015
|
|
4WWN
 
 | | Crystal structure of human PI3K-gamma in complex with (S)-N-(1-(7-fluoro-2-(pyridin-2-yl)quinolin-3-yl)ethyl)-9H-purin-6-amine AMG319 inhibitor | | 分子名称: | N-{(1S)-1-[7-fluoro-2-(pyridin-2-yl)quinolin-3-yl]ethyl}-9H-purin-6-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | 著者 | Whittington, D.A, Tang, J, Yakowec, P. | | 登録日 | 2014-11-11 | | 公開日 | 2014-12-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery and in Vivo Evaluation of (S)-N-(1-(7-Fluoro-2-(pyridin-2-yl)quinolin-3-yl)ethyl)-9H-purin-6-amine (AMG319) and Related PI3K delta Inhibitors for Inflammation and Autoimmune Disease.
J.Med.Chem., 58, 2015
|
|
4WXJ
 
 | |
6XMX
 
 | | Cryo-EM structure of BCL6 bound to BI-3802 | | 分子名称: | 2-[6-[[5-chloranyl-2-[(3~{S},5~{R})-3,5-dimethylpiperidin-1-yl]pyrimidin-4-yl]amino]-1-methyl-2-oxidanylidene-quinolin-3-yl]oxy-~{N}-methyl-ethanamide, B-cell lymphoma 6 protein | | 著者 | Yoon, H, Burman, S.S.R, Hunkeler, M, Nowak, R.P, Fischer, E.S. | | 登録日 | 2020-07-01 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Small-molecule-induced polymerization triggers degradation of BCL6.
Nature, 588, 2020
|
|
7QR9
 
 | | Crystal structure of CK1 delta in complex with PK-09-82 | | 分子名称: | 1,2-ETHANEDIOL, 4-[5-(4-fluorophenyl)-3-(pyridin-4-ylmethyl)imidazol-4-yl]-1~{H}-pyrrolo[2,3-b]pyridine, Casein kinase I isoform delta, ... | | 著者 | Chaikuad, A, Khirsariya, P, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2022-01-10 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery of Potent and Exquisitely Selective Inhibitors of Kinase CK1 with Tunable Isoform Selectivity.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7QRB
 
 | | Crystal structure of CK1 delta in complex with PK-09-129 | | 分子名称: | 3-(dimethylamino)-~{N}-[4-[4-(4-fluorophenyl)-5-(1~{H}-pyrrolo[2,3-b]pyridin-4-yl)imidazol-1-yl]cyclohexyl]propane-1-sulfonamide, Casein kinase I isoform delta, SULFATE ION | | 著者 | Chaikuad, A, Khirsariya, P, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2022-01-10 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Discovery of Potent and Exquisitely Selective Inhibitors of Kinase CK1 with Tunable Isoform Selectivity.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4X2J
 
 | | Selection of fragments for kinase inhibitor design: decoration is key | | 分子名称: | 4-[(3-aminophenyl)amino]pyrido[2,3-d]pyrimidin-5(8H)-one, SULFATE ION, TGF-beta receptor type-1 | | 著者 | Czodrowski, P, Hoelzemann, G, Barnickel, G, Greiner, H, Musil, D. | | 登録日 | 2014-11-26 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Selection of fragments for kinase inhibitor design: decoration is key.
J.Med.Chem., 58, 2015
|
|
