5V6I
 
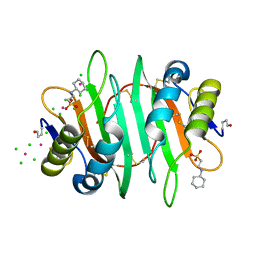 | | Glycan binding protein Y3 from mushroom Coprinus comatus possesses anti-leukemic activity - Pt derivative | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, PLATINUM (II) ION, ... | | Authors: | Li, K, Zhang, P, Gang, Y, Xia, C, Polston, J.E, Li, G, Li, S, Lin, Z, Yang, L.-J, Bruner, S.D, Ding, Y. | | Deposit date: | 2017-03-16 | | Release date: | 2017-08-16 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cytotoxic protein from the mushroom Coprinus comatus possesses a unique mode for glycan binding and specificity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1CIM
 
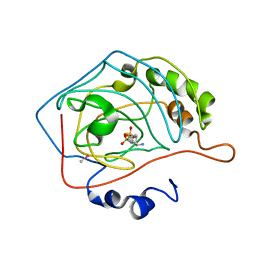 | | THE POSITIONS OF HIS-64 AND A BOUND WATER IN HUMAN CARBONIC ANHYDRASE II UPON BINDING THREE STRUCTURALLY RELATED INHIBITORS | | Descriptor: | (4S-TRANS)-4-(AMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO (2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, CARBONIC ANHYDRASE II, METHYL MERCURY ION, ... | | Authors: | Smith, G.M, Alexander, R.S, Christianson, D.W, Mckeever, B.M, Ponticello, G.S, Springer, J.P, Randall, W.C, Baldwin, J.J, Habecker, C.N. | | Deposit date: | 1993-10-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Positions of His-64 and a bound water in human carbonic anhydrase II upon binding three structurally related inhibitors.
Protein Sci., 3, 1994
|
|
5V3A
 
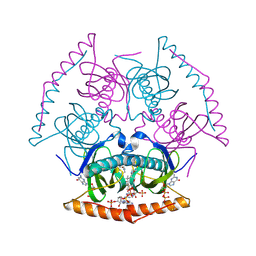 | |
2E98
 
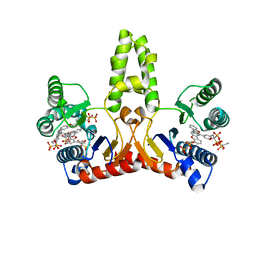 | | E. coli undecaprenyl pyrophosphate synthase in complex with BPH-629 | | Descriptor: | Undecaprenyl pyrophosphate synthetase, [2-(3-DIBENZOFURAN-4-YL-PHENYL)-1-HYDROXY-1-PHOSPHONO-ETHYL]-PHOSPHONIC ACID | | Authors: | Guo, R.T, Ko, T.P, Cao, R, Liang, P.H, Oldfield, E, Wang, A.H.J. | | Deposit date: | 2007-01-24 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5N0C
 
 | | Crystal structure of the tetanus neurotoxin in complex with GM1a | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tetanus toxin, ZINC ION, ... | | Authors: | Masuyer, G, Conrad, J, Stenmark, P. | | Deposit date: | 2017-02-02 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of the tetanus toxin reveals pH-mediated domain dynamics.
EMBO Rep., 18, 2017
|
|
4ALI
 
 | | Crystal structure of S. aureus FabI in complex with NADP and triclosan (P1) | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Kisker, C. | | Deposit date: | 2012-03-04 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
2RU3
 
 | | Solution structure of c.elegans SUP-12 RRM in complex with RNA | | Descriptor: | Protein SUP-12, isoform a, RNA (5'-R(*GP*UP*GP*UP*GP*C)-3') | | Authors: | Takahashi, M, Kuwasako, K, Unzai, S, Tsuda, K, Yoshikawa, S, He, F, Kobayashi, N, Guntert, P, Shirouzu, M, Ito, T, Tanaka, A, Yokoyama, S, Hagiwara, M, Kuroyanagi, H, Muto, Y. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RBFOX and SUP-12 sandwich a G base to cooperatively regulate tissue-specific splicing
Nat.Struct.Mol.Biol., 21, 2014
|
|
5N49
 
 | | BRPF2 in complex with Compound 7 | | Descriptor: | 2-(1,3,6-trimethyl-2-oxidanylidene-benzimidazol-5-yl)benzo[de]isoquinoline-1,3-dione, Bromodomain-containing protein 1 | | Authors: | Bouche, L, Christ, C.D, Siegel, S, Fernandez-Montalvan, A.E, Holton, S.J, Fedorov, O, ter Laak, A, Sugawara, T, Stoeckigt, D, Tallant, C, Bennett, J, Monteiro, O, Saez, L.D, Siejka, P, Meier, J, Puetter, V, Weiske, J, Mueller, S, Huber, K.V.M, Hartung, I.V, Haendler, B. | | Deposit date: | 2017-02-10 | | Release date: | 2017-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Benzoisoquinolinediones as Potent and Selective Inhibitors of BRPF2 and TAF1/TAF1L Bromodomains.
J. Med. Chem., 60, 2017
|
|
1JR2
 
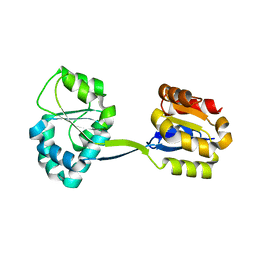 | | Structure of Uroporphyrinogen III Synthase | | Descriptor: | UROPORPHYRINOGEN-III SYNTHASE | | Authors: | Mathews, M.A, Schubert, H.L, Whitby, F.G, Alexander, K.J, Schadick, K, Bergonia, H.A, Phillips, J.D, Hill, C.P. | | Deposit date: | 2001-08-10 | | Release date: | 2001-11-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of human uroporphyrinogen III synthase.
EMBO J., 20, 2001
|
|
3BBD
 
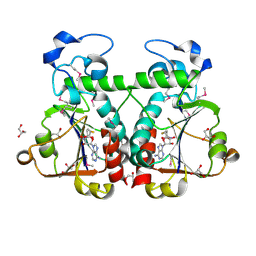 | | M. jannaschii Nep1 complexed with S-adenosyl-homocysteine | | Descriptor: | GLYCEROL, Ribosome biogenesis protein NEP1-like, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Taylor, A.B, Meyer, B, Leal, B.Z, Kotter, P, Hart, P.J, Entian, K.-D, Wohnert, J. | | Deposit date: | 2007-11-09 | | Release date: | 2008-02-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site.
Nucleic Acids Res., 36, 2008
|
|
1PQV
 
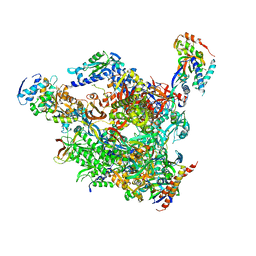 | | RNA polymerase II-TFIIS complex | | Descriptor: | DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, ... | | Authors: | Kettenberger, H, Armache, K.-J, Cramer, P. | | Deposit date: | 2003-06-19 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Architecture of the RNA Polymerase II-TFIIS Complex and Implications for mRNA Cleavage
Cell(Cambridge,Mass.), 114, 2003
|
|
3UOV
 
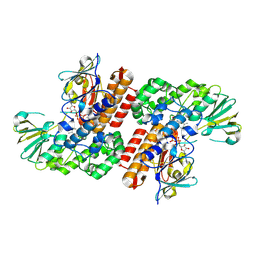 | | Crystal Structure of OTEMO (FAD bound form 1) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.045 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
2RU5
 
 | |
3UP5
 
 | | Crystal Structure of OTEMO complex with FAD and NADP (form 4) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3BEW
 
 | | 10mer Crystal Structure of chicken MHC class I haplotype B21 | | Descriptor: | 10-mer from Tubulin beta-6 chain, Beta-2-microglobulin, Major histocompatibility complex class I glycoprotein haplotype B21 | | Authors: | Koch, M, Camp, S, Collen, T, Avila, D, Salomonsen, J, Wallny, H.J, van Hateren, A, Hunt, L, Jacob, J.P, Johnston, F, Marston, D.A, Shaw, I, Dunbar, P.R, Cerundolo, V, Jones, E.Y, Kaufman, J. | | Deposit date: | 2007-11-20 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of an MHC class I molecule from b21 chickens illustrate promiscuous Peptide binding
Immunity, 27, 2007
|
|
5UR0
 
 | | Crystallographic structure of glyceraldehyde-3-phosphate dehydrogenase from Naegleria gruberi | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Machado, A.T.P, Silva, M, Iulek, J. | | Deposit date: | 2017-02-09 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural studies of glyceraldehyde-3-phosphate dehydrogenase from Naegleria gruberi, the first one from phylum Percolozoa.
Biochim. Biophys. Acta, 1866, 2018
|
|
7EZV
 
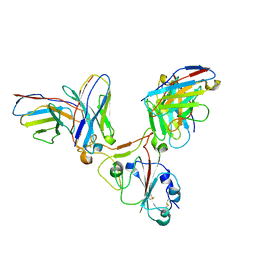 | |
5N41
 
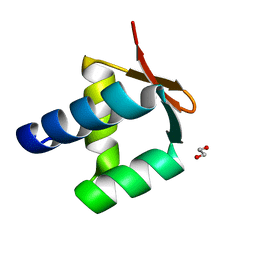 | | Archaeal DNA polymerase holoenzyme - SSO6202 at 1.35 Ang resolution | | Descriptor: | 1,2-ETHANEDIOL, PolB1 Binding Protein 2 | | Authors: | Yan, J, Beattie, T.R, Rojas, A.L, Schermerhorn, K, Gristwood, T, Trinidad, J.C, Albers, S.V, Roversi, P, Gardner, A.F, Abrescia, N.G.A, Bell, S.D. | | Deposit date: | 2017-02-09 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Identification and characterization of a heterotrimeric archaeal DNA polymerase holoenzyme.
Nat Commun, 8, 2017
|
|
3G5V
 
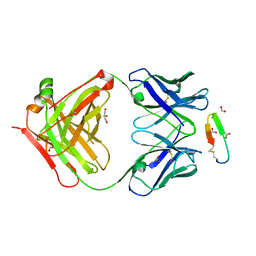 | | Antibodies Specifically Targeting a Locally Misfolded Region of Tumor Associated EGFR | | Descriptor: | 806 light chain, 808 heavy chain, ACETATE ION, ... | | Authors: | Garrett, T.P.J, Burgess, A.W, Huyton, T, Xu, Y. | | Deposit date: | 2009-02-05 | | Release date: | 2010-02-09 | | Last modified: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Antibodies specifically targeting a locally misfolded region of tumor associated EGFR
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7EY4
 
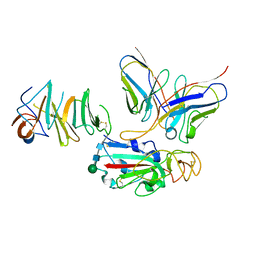 | | Local CryoEM of the SARS-CoV-2 S6PV2 in complex with BD-667 | | Descriptor: | BD-667 H, BD-667 L, Spike glycoprotein, ... | | Authors: | Liu, P.L. | | Deposit date: | 2021-05-29 | | Release date: | 2021-09-08 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structures of SARS-CoV-2 B.1.351 neutralizing antibodies provide insights into cocktail design against concerning variants.
Cell Res., 31, 2021
|
|
1PQQ
 
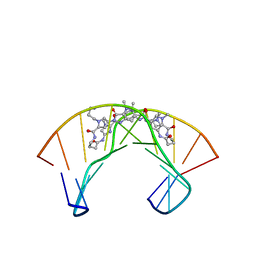 | | NMR Structure of a Cyclic Polyamide-DNA Complex | | Descriptor: | 45-(3-AMINOPROPYL)-5,11,22,28,34-PENTAMETHYL-3,9,15,20,26,32,38,43-OCTAOXO-2,5,8,14,19,22,25,28,31,34,37,42,45,48-TETRADECAAZA-11-AZONIAHEPTACYCLO[42.2.1.1~4,7~.1~10,13~.1~21,24~.1~27,30~.1~33,36~]DOPENTACONTA-1(46),4(52),6,10(51),12,21(50),23,27(49),29,33(48),35,44(47)-DODECAENE, 5'-D(*CP*GP*CP*TP*AP*AP*CP*AP*GP*GP*C)-3', 5'-D(*GP*CP*CP*TP*GP*TP*TP*AP*GP*CP*G)-3' | | Authors: | Zhang, Q, Dwyer, T.J, Tsui, V, Case, D.A, Cho, J, Dervan, P.B, Wemmer, D.E. | | Deposit date: | 2003-06-18 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Cyclic Polyamide-DNA Complex.
J.Am.Chem.Soc., 126, 2004
|
|
3QPL
 
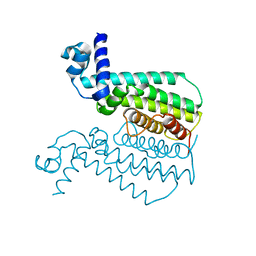 | | G106W mutant of EthR from Mycobacterium tuberculosis | | Descriptor: | HTH-type transcriptional regulator EthR | | Authors: | Carette, X, Willery, E, Hoos, S, Lecat-Guillet, N, Frenois, F, Dirie, B, Villeret, V, England, P, Deprez, B, Locht, C, Willand, N, Baulard, A. | | Deposit date: | 2011-02-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural activation of the transcriptional repressor EthR from Mycobacterium tuberculosis by single amino acid change mimicking natural and synthetic ligands.
Nucleic Acids Res., 40, 2012
|
|
3UU0
 
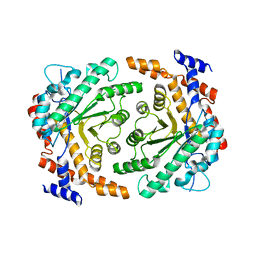 | | Crystal structure of L-rhamnose isomerase from Bacillus halodurans in complex with Mn | | Descriptor: | L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Doan, T.T.N, Prabhu, P, Kim, J.K, Jeya, M, Kang, L.W, Lee, J.K. | | Deposit date: | 2011-11-27 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based studies on the metal binding of two-metal-dependent sugar isomerases.
Febs J., 281, 2014
|
|
2NY7
 
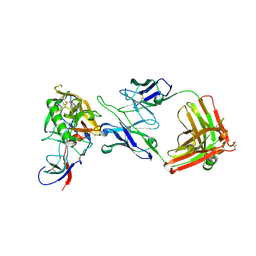 | | HIV-1 gp120 Envelope Glycoprotein Complexed with the Broadly Neutralizing CD4-Binding-Site Antibody b12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY b12, HEAVY CHAIN, ... | | Authors: | Zhou, T, Xu, L, Dey, B, Hessell, A.J, Van Ryk, D, Xiang, S.H, Yang, X, Zhang, M.Y, Zwick, M.B, Arthos, J, Burton, D.R, Dimitrov, D.S, Sodroski, J, Wyatt, R, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2006-11-20 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural definition of a conserved neutralization epitope on HIV-1 gp120.
Nature, 445, 2007
|
|
4DJZ
 
 | | Catalytic fragment of masp-1 in complex with its specific inhibitor developed by directed evolution on sgci scaffold | | Descriptor: | Mannan-binding lectin serine protease 1 heavy chain, Mannan-binding lectin serine protease 1 light chain, Protease inhibitor SGPI-2 | | Authors: | Heja, D, Harmat, V, Fodor, K, Wilmanns, M, Dobo, J, Kekesi, K.A, Zavodszky, P, Gal, P, Pal, G. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Monospecific Inhibitors Show That Both Mannan-binding Lectin-associated Serine Protease-1 (MASP-1) and -2 Are Essential for Lectin Pathway Activation and Reveal Structural Plasticity of MASP-2.
J.Biol.Chem., 287, 2012
|
|
